
*** The following text is out-of-date.***
For the latest news about COVID-19, please open the COVID Reference homepage.
By Bernd Sebastian Kamps
Copy-editor: Rob Camp
Published 31 January | Revised 3 April
Summary Week 13
3 April
Previous weeks 04 | 05 | 06 | 07 | 08 | 09 | 11 | 12
See the full chapter after this week’s summary.
B.1.1.7 in the US
B.1.1.7 is now spreading throughout the US. In some states, the impressive US vaccination campaign will help control the B.1.1.7-driven epidemic in the epidemic, while other states will see important surges of new cases (Figure S1). These differences in outcome will later help evaluate the usefulness of mitigation measures (Figure S2).
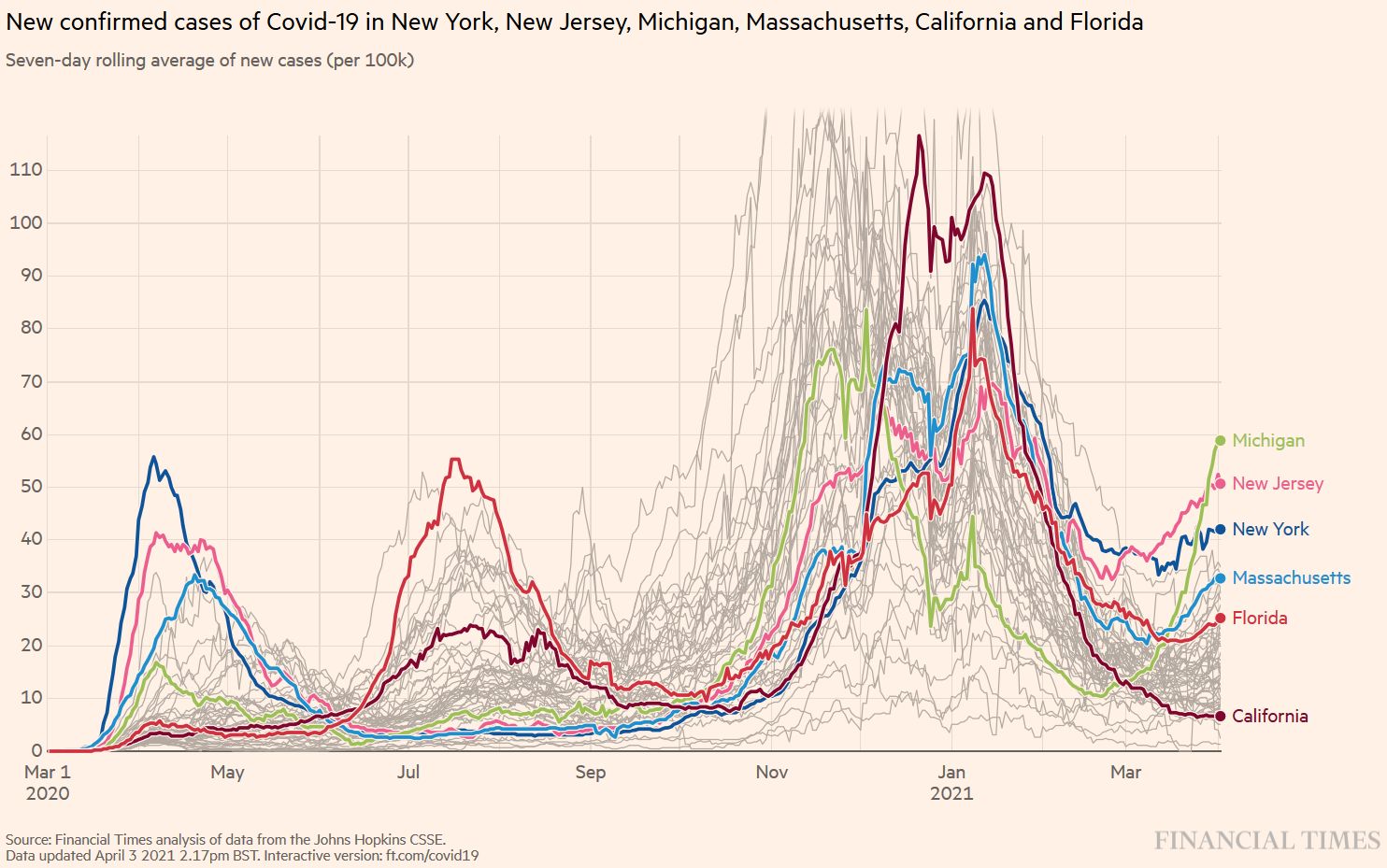
Figure S1. B.1.1.7 Spring wave in selected US states. Source and copyright: Financial Times 2021, accessed 3 April.
Figure S2. How to get out of the pandemic. Source and copyright: Wiley Miller, https://www.gocomics.com/nonsequitur, accessed 3 April.
B.1.1.7 vs B.1.351
Which variants – B.1.1.7 (first detected in England), B.1.351 (South Africa), or P.1 (Brazil) – will ultimately prevail? For now, B.1.1.7 seems to be in the lead. Data from the French Moselle department shows that in early February, the two variants from the southern hemisphere B.1.351 and P.1. accounted for 40% of new infections, compared to only 20% for B.1.1.7. One month later, B.1.1.7 was found in almost 50% of new cases (Figure S3). How long will this dominance last? Nobody knows. Once a majority of the population has been vaccinated in early autumn, the variant cards will be reshuffled.

Figure S3. Take-over of the English variant B.1.1.7 in the French Moselle department. (Souche classique: historical lineage) Source and copyright: Herzberg N. Covid-19 : qui va gagner la bataille des variants? Le Monde 2021, published 31 March. Full text: https://www.lemonde.fr/planete/article/2021/03/31/covid-19-qui-va-gagner-la-bataille-des-variants_6075043_3244.html
Important publications of the week
Transmissibility
Peng J, Liu J, Mann SA, et al. Estimation of secondary household attack rates for emergent spike L452R SARS-CoV-2 variants detected by genomic surveillance at a community-based testing site in San Francisco. Clinical Infectious Diseases 31 March 2021, ciab283, https://academic.oup.com/cid/advance-article/doi/10.1093/cid/ciab283/6206738?searchresult=1
The new lineages B.1.427 and B.1.429 (the “California” or “West Coast” variants) share S gene non-synonymous mutations at sites 13, 152, 452, and 614 and were seen during the December 2020 to February 2021 period when California was experiencing a huge peak. Whereas no instances of B.1.1.7, or independent N501Y mutations were detected in the sample population, the authors found a modest transmissibility increase of the West Coast variants. Household contacts exposed to these variants were at higher risk of infection compared to those exposed to lineages lacking these variants (0.36 vs 0.29, RR = 1.28; 95% CI: 1.00-1.64). Ct values did not differ.
Immune evasion
Dejnirattisai W, Zhou D, Supasa P, et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell March 30, 2021. https://www.cell.com/cell/fulltext/S0092-8674(21)00428-1
All new strains (P.1 from Brazil, B.1.351 from South Africa and B.1.1.7 from the UK) have mutations in the ACE2 binding site with P.1 and B.1.351 having a virtually identical triplet: E484K, K417N/T and N501Y, conferring similar increased affinity for ACE2. Surprisingly, P.1 was significantly less resistant to naturally acquired or vaccine induced antibody responses than B.1.351, suggesting that changes outside the receptor-binding domain impact neutralization.
Ikegame S, Siddiquey MNA, Hung CT, et al. Qualitatively distinct modes of Sputnik V vaccine-neutralization escape by SARS-CoV-2 Spike variants. medRxiv 2021, posted 2 April. Full text: https://doi.org/10.1101/2021.03.31.21254660
The importance of the 2-P (proline) substitution in the spike protein in vaccines.
Vaccine efficacy
Emary KR, Gulobchik T, Aley PK. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet March 30, 2021. https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(21)00628-0/fulltext
This post-hoc analysis of the efficacy of the adenoviral vector vaccine, ChAdOx1 nCoV-19 (AZD1222), revealed that laboratory virus neutralization activity by vaccine-induced antibodies was lower against B.1.1.7. However, clinical vaccine efficacy against symptomatic NAAT positive infection was good, with 70% (95% CI 44–85) for B.1.1.7 and 82% (68–89) for other lineages.
B.1.1.7
Washington NL, Gangavarapu K, Zeller M, et al. Genomic epidemiology identifies emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell 2021, 30 March. Full-text: https://www.cell.com/cell/fulltext/S0092-8674(21)00383-4
Kristian Andersen, Nicole L. Washington, Karthik Gangavarapu and colleagues from Helix and the Scripps Research Institute, La Jolla, sequenced 212 B.1.1.7 genomes collected in the U.S. from December 2020 to January 2021. They found a doubling rate of a little over a week and an increased transmission rate of 35-45%. The authors showed in February that the U.S. was on a similar trajectory as other countries where B117 rapidly became the dominant SARS-CoV-2 variant and warn that immediate and decisive action to minimize COVID-19 morbidity and mortality.
P.1
Francisco MA, Zavascki AP, Lamb WP, et al. Detection of SARS-CoV-2 lineage P.1 in patients from a region with exponentially increasing hospitalisation rate, February 2021, Rio Grande do Sul, Southern Brazil. Euro Surveill Accepted: 25 Mar 2021, 2021. Full text: https://doi.org/10.2807/1560-7917.ES.2021.26.12.2100276
From epidemiological week 6, starting on 7 February 2021, until 6 March, the number of hospitalizations for COVID-19 in Rio Grande do Sul, the southernmost state of Brazil in the South region, increased from 1738 inpatients to 6995 (3.8-fold). This resulted in the collapse of the state healthcare system. The overwhelming increase in hospitalizations temporally coincided with the finding that lineage P.1 became predominant (although a small number of specimens was taken).
Peer-reviewed former preprints
Cele S, Gazy I, Jackson L, et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021 Mar 29. PubMed: https://pubmed.gov/33780970. Full-text: https://doi.org/10.1038/s41586-021-03471-w
We already knew that people previously infected with the non-B.1.351 variant don’t neutralize B.1.351 very effectively. Now Alex Sigal, Tulio de Oliveira, Sandile Cele and colleagues show that people infected with B.1.351 can neutralize both B.1.351 and (to a slightly lesser extent) ‘regular’ non-B.1.351 viruses (Cele 2021). If these data are confirmed, a variant B.1.351-targeted booster vaccine could be a solution for countries where B.1.351 is the dominant strain.
Previous Weekly Summaries
Please find the previous summaries at
- 27 March: https://covidreference.com/variants-27-march
- 20 March: https://covidreference.com/variants-20-march
- 6 March: https://covidreference.com/variants-6-march
- 27 February: https://covidreference.com/variants-27-february
- 20 February: https://covidreference.com/variants-20-february
- 13 February: https://covidreference.com/variants-13-february
- 6 February: https://covidreference.com/variants-6-february
- 31 January: https://covidreference.com/variants-31-january
Introduction
With viruses, some mutations emerge while others recede. Rarely does one or more mutations confer a “selective advantage” to a new variant, for example enhanced transmissibility. When it does happen, such variants can then become the new dominant virus.
Over the last three months, several new SARS-CoV-2 variants have been described that are more transmissible, may escape both natural and vaccine-induced immunity and could impact COVID-19 morbidity and mortality. They are creating a new pandemic within the pandemic. In countries like England, South Africa, Brazil, Ireland, Portugal and Israel, they have clearly modified the dynamic of the latest outbreaks for the worse. More transmissible SARS-CoV-2 variants will replace older variants – everywhere! Countries where the prevalence of these new variants is still low should anticipate rapid spread within the next weeks and months and plan ahead accordingly.
The current trio infernale:
- 1.1.7 (first described in England; Rambaut 2020)
- 1.351 (first described in South Africa; Tegally 2021)
- P.1 (first described in Brazil; Faria 2021)
Of note, although these variants evolved independently in different places around the globe, they share key mutations which are involved in receptor binding. This viral evolution is a normal process known from seasonal coronaviruses (Wong AHM 2017, Eguia 2020, Kistler 2021) and has recently been reproduced in vitro (Zahradnik 2020). Convergent evolution suggests that under the pressure of an increasing number of people having developed antibodies against SARS-CoV-2, the virus is developing a more perfect configuration.
The variant mutations may affect the COVID-19 pandemic in multiple ways:
- Increased transmissibility
- Increased severity of illness
- Diminished protection from previous SARS-CoV-2 infection
- Diminished response to vaccines
- Diminished response to monoclonal antibodies
A higher rate of transmission will lead to more COVID-19 cases, increase the number of persons who need clinical care, exacerbate the burden on an already strained health care system, and finally result in more deaths (Galloway 2020). The increased transmissibility of new variants may therefore require an even more rigorous implementation of vaccination and mitigation measures (e.g., distancing, masking, and hand hygiene) to control the spread of SARS-CoV-2. These measures will be more effective if they are instituted sooner rather (Figure 3.)
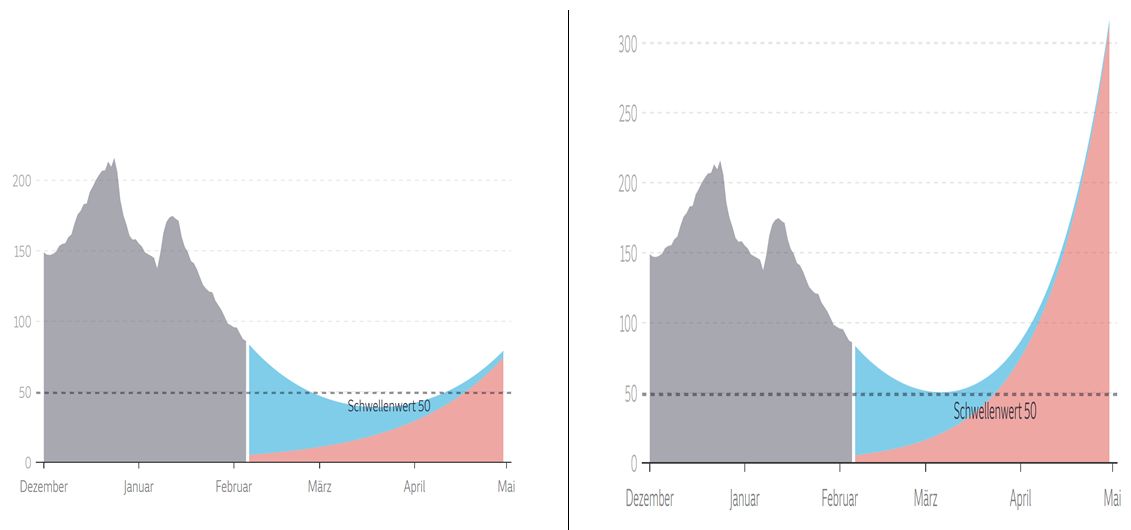
Figure 3. March to May 2021: possible scenarios for national epidemics. Blue: new SARS-CoV-2 infections due to old variants; red: new SARS-CoV-2 infections due to new variants. On the left, mitigation measures succeed in keeping case numbers low until the general availability of vaccines for the adult population. On the left, an uncontrolled epidemic is fuelled by the higher transmissibility of the new SARS-CoV-2 variants. Graphic copyright: Süddeutsche Zeitung, 5 February (Berndt 2021).
Epidemiology
B.1.1.7 is now the new ‘normalcy’ in most European countries. It is about 50% more transmissible than historical strains and is expected to increase the basic reproductive number R0 by 0.3 to 0.4. In regions where R0 is only slightly below 1, a new pandemic wave should be anticipated. Recent studies suggest that people infected with B.1.1.7 may have a more than 50% higher risk of hospitalisation (Bager 2021) and death (NERVTAG 20210211, Davies 2021). B.1.1.7 is on its way to become the dominant global strain.
In most countries, COVID-19 vaccines will have no impact on the current B.1.1.7 wave. Countries that are unable to vaccinate the adult population within the next weeks will see a surge of COVID-19 cases, annihilating every prospect of a ‘normal’ Easter vacation. Countries that are unable to vaccinate their population before June will not see a normal summer season either. Finally, countries that are unable to vaccinate well over 80% of their population (vaccine skepticism, limited access, etc.) will struggle to return to pre-COVID-19 normalcy.
The following curves describe the “One month – 20 to 80” rule of the B.1.1.7 ‘take-over’ (Figures 4 and 5). Within one month, the percentage of the B.1.1.7 variant progresses from 20% to 80% in a given population. These so-called Wenseleers curves are helpful for predicting an imminent recrudescence of the COVID-19 pandemic in regions and countries where B.1.1.7 is not yet dominant.
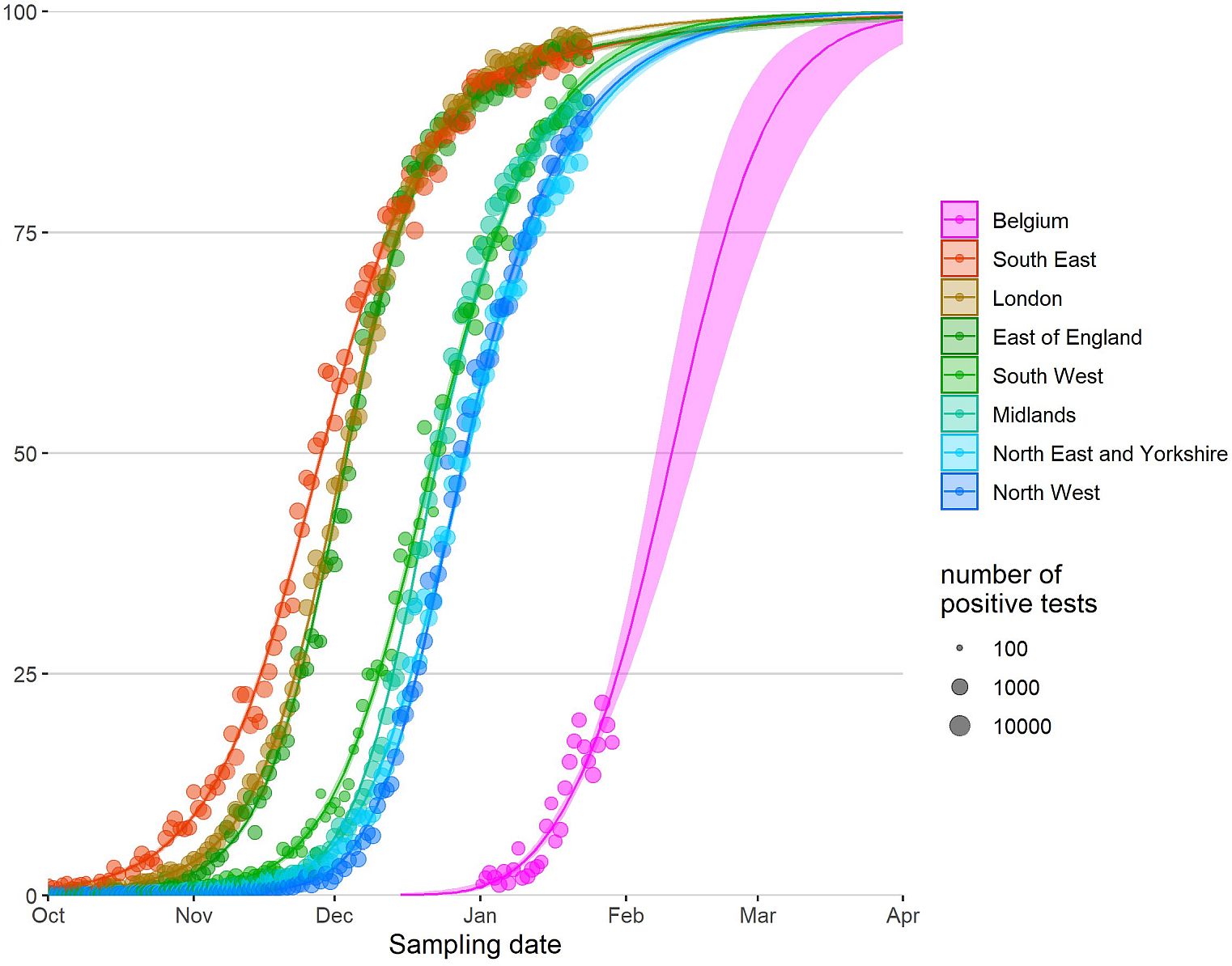
Figure 4. Relative abundance of B.1.1.7 in England and Belgium, posted on 3 February by Tom Wenseleers, https://bit.ly/3pJP7Db. The data show that the B.1.1.7 variant progressed from 20% to 80% of all circulating strains within about four weeks. Other European countries have since followed the same steep pattern.
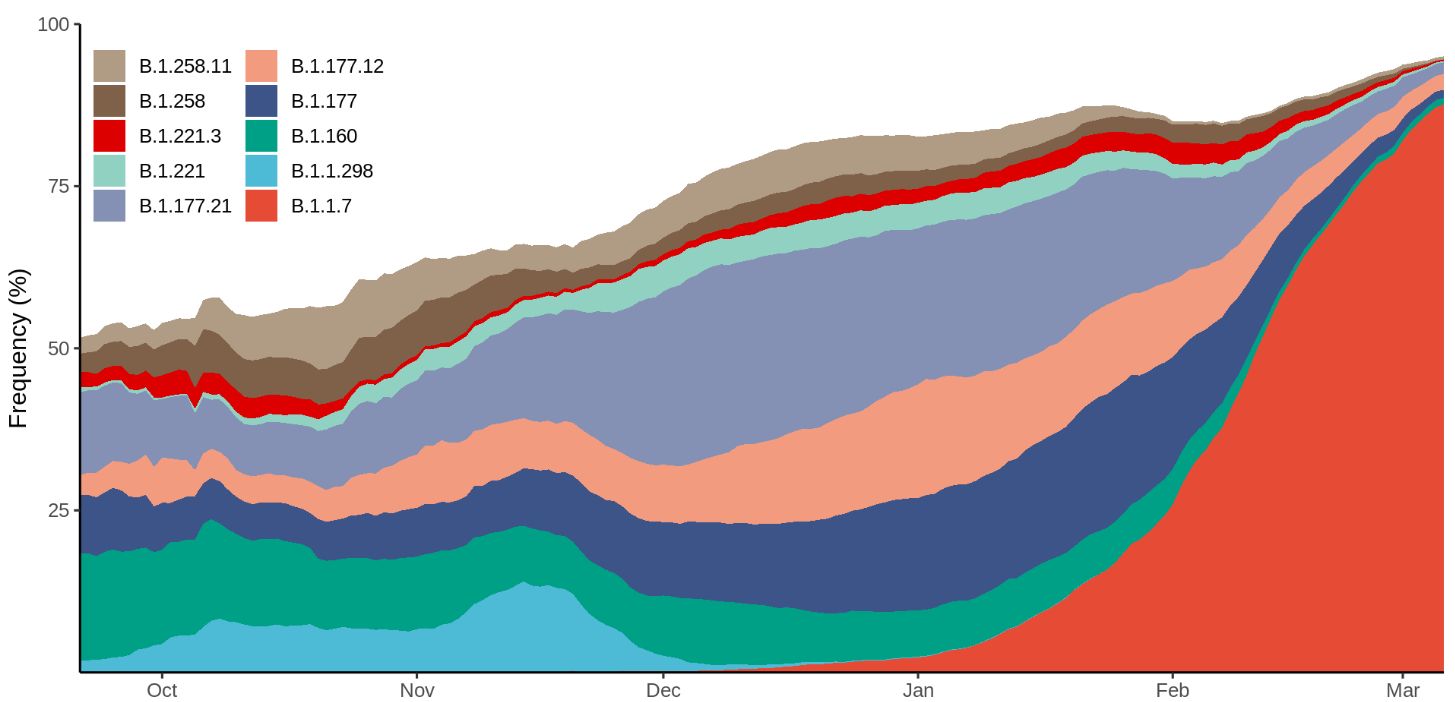
Figure 5. Frequency of the 10 most abundant lineages across Denmark within the last 6 months. The frequency is calculated as a centered 14-day rolling average. Weekly increase: 2, 4, 7, 13, 20, 30, 47, 66, 75, 85, 93%. Source: Danish Covid-19 Genome Consortium, 17 March 2021, https://www.covid19genomics.dk/statistics. Accessed 19 March.
Once the proportion of B.1.1.7 of newly diagnosed SARS-CoV-2 cases exceeds 80%, the epidemic activity intensifies within 2 to 4 weeks. Figure 6 shows the daily new confirmed COVID-19 cases in selected European countries. The increased transmissibility of B.1.1.7 leads to an increased number of infections in several countries, which in turn leads to higher hospitalization and death rates.
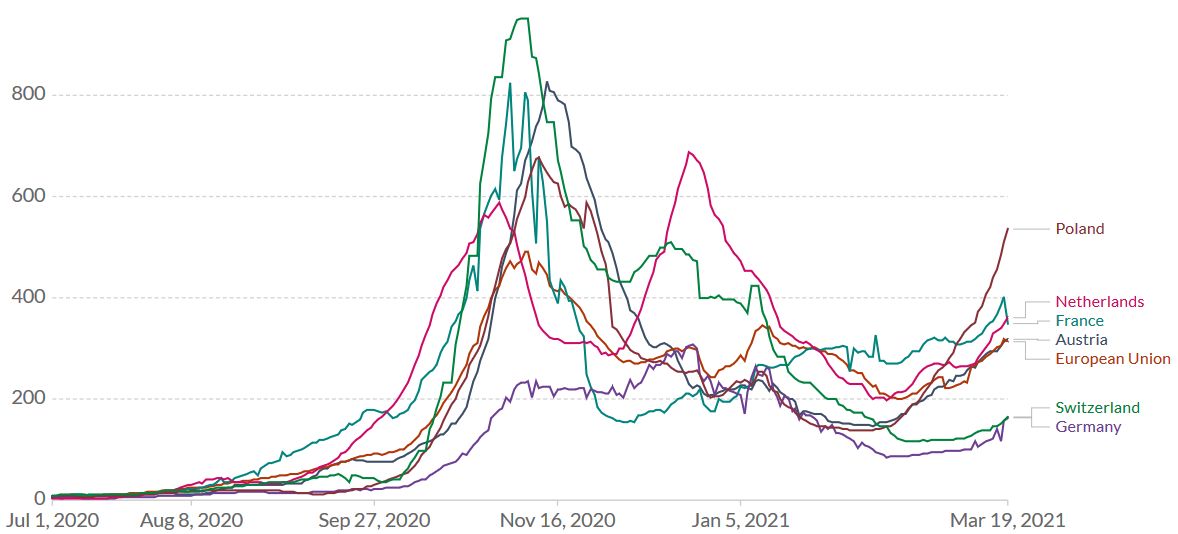
Figure 6. The English B.1.1.7 variant has reached Continental Europe. Shown is the rolling 7-day average per million people for selected European countries. To recalculate these values as the ‘cumulative 7-day rolling incidence per 100.000 people’, multiply the values by 0.7. Example: the March 19 value for the European Union, 313, is equivalent to a cumulative 7-day incidence of 219/100.000 people. Source and copyright: Our World in Data – Johns Hopkins University CSSE COVID-19 Data, accessed 20 March.
A recent modelling study suggests that vaccination alone might be insufficient to contain the SARS-CoV-2 pandemic (Moore 2021; see also the comment by Contreras & Priesemann 2021). The authors assume a default vaccine uptake of 95% in those aged 80 years and older, 85% in those aged 50–79 years, and 75% in those aged 18–49 years. Even with the optimistic assumption that the vaccine will prevent 85% of infections, the authors estimate R to be 1.58 (95% CI: 1.36–1.84) once all eligible adults have been offered both doses of the vaccine. Tough times on the horizon.
United Kingdom
In the UK, the first case of the B.1.1.7 variant (ie, 20B/501Y.V1 or VOC 202012/01) was retrospectively dated to 20 September. In Kent, a county in South East England, cases continued to increase during a lockdown in November, despite having the same restrictions as other regions. When, on 2 December, England lifted its lockdown, the proportion in England of B.1.1.7 continued to increase sharply in Kent and then rapidly in Greater London and other parts of the southeast (Kirby 2021), rising to over 70% at the beginning of January 2021. Areas with the highest B.1.1.7 incidence coincided with areas reporting higher levels of patient hospitalisation (Gravagnuolo 2021). In England, B.1.1.7 took around 6 weeks to go from less than 20% of cases to over 80% (Public Health England 20210126, Figure 7).
The good news from the UK: lockdowns are efficient against the SARS-CoV-2 variant too. After a peak of the rolling 7-day average on 9 January, the numbers started decreasing (Figure 8). Whether the spike protein mutation E484K which was detected in eleven B.1.1.7 sequences at the end of January 2021 (Public Health England 20210126) will eventually spread more widely, is unknown.
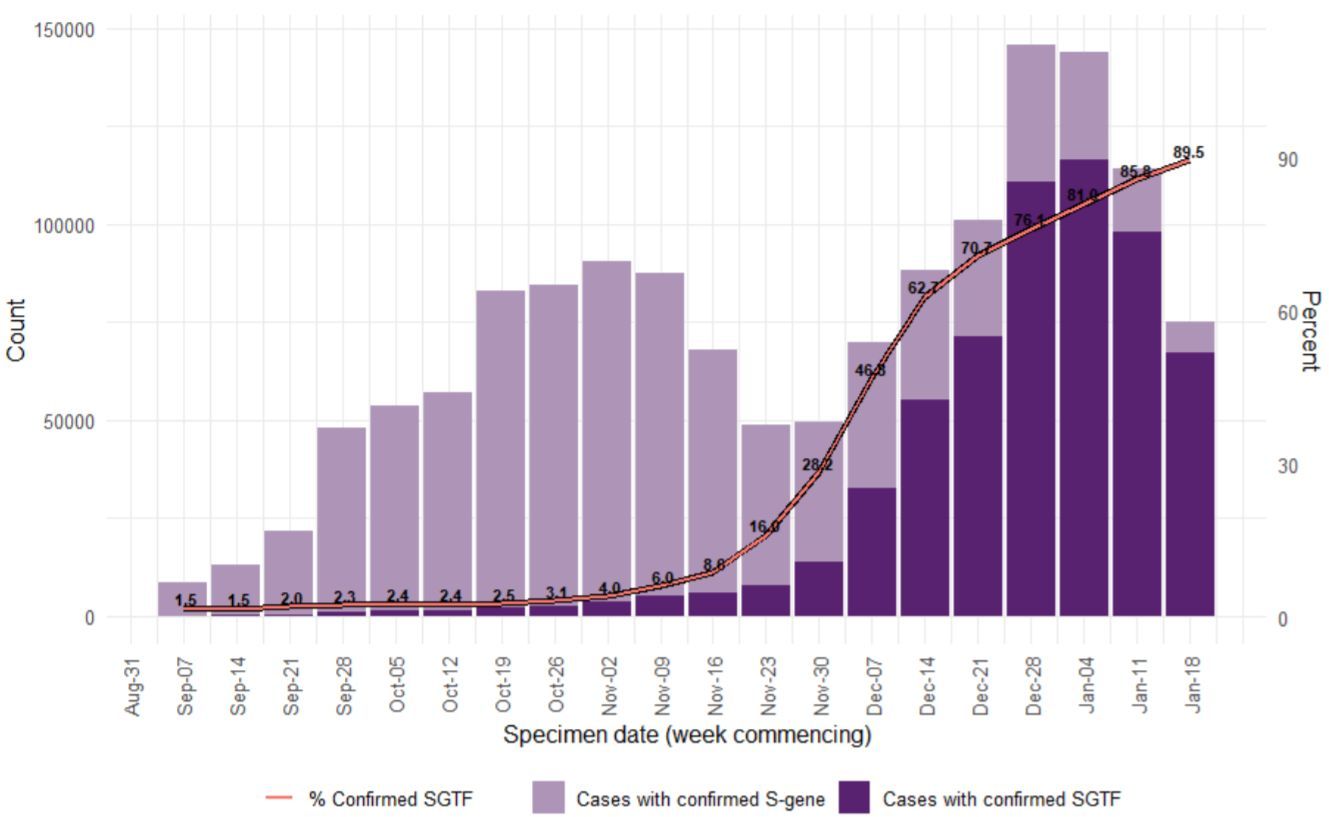
Figure 7. Weekly number (bars) and proportion (line) of B.1.1.7 (SGTF: “S–gene target failure”; deep purple) COVID-19 cases (7 September 2020 to 24 January 2021) (Public Health England 20210126).
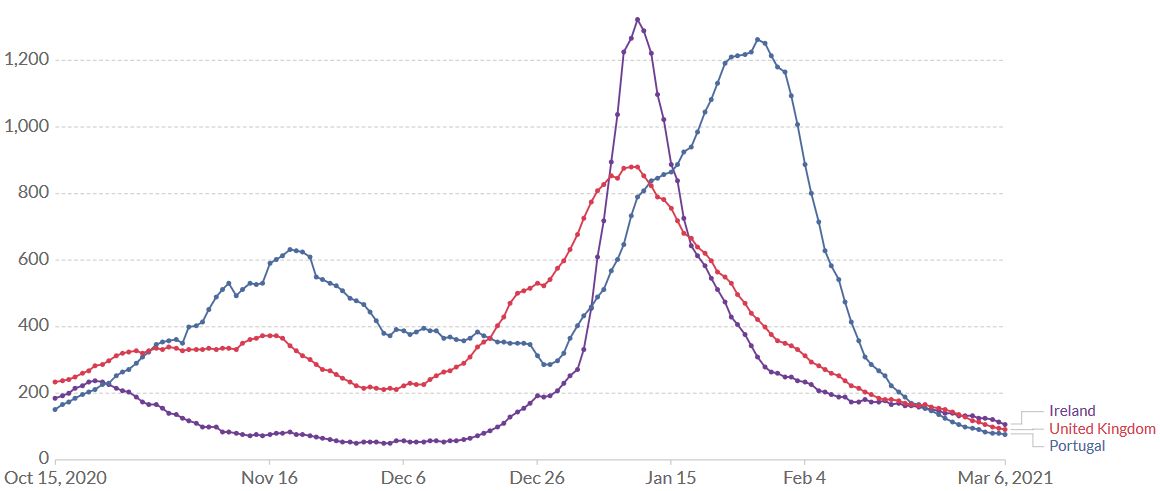
Figure 8. United Kingdom, Ireland, Portugal, 6 March. Lockdowns are efficient against B.1.1.7. After a peak of the rolling 7-day average on 10 January (UK, Ireland) and 28 January (Portugal), the number of daily new confirmed SARS-CoV-2 cases decreased rapidly. Source: Our World in Data – Johns Hopkins University CSSE COVID-19 Data.
South Africa
In South Africa, B.1.351 (also called 501Y.V2) was first detected in early October 2020 and is now the most prevalent variant in the country. Like B.1.1.7, it has an increased transmissibility. After a peak of the rolling 7-day average on 11 January, the numbers in South Africa decreased rapidly (Figure 2).
As of 20 March, B.1.351 has been identified in 48 countries. Clusters of this variant are currently being investigated in France and Austria (Tyrol). Israel and the UK have also reported cases or clusters of non-travel-related B.1.351 cases (ECDC 20210121). Lockdowns are efficient against B.1.351. Find more information about B.1.351 on page 36.
Brazil
In January 2021, the P.1 variant was identified in 42% (13 out of 31) of RT-PCR positive samples collected between 15 and 23 December in Manaus, Amazonia, Brazil (Faria 2021). At the time, Manaus was experiencing an upsurge in COVID-19 cases. P.1 is refractory to multiple neutralizing monoclonal antibodies and more resistant to neutralization by (first-wave) convalescent plasma (de Souza 2021, Wang P 2021). Five months after booster immunization with the Chinese CoronaVac vaccine, plasma from vaccinated individuals failed to efficiently neutralize P.1 lineage isolates (de Souza 2021). In a pre-print paper, P.1 has been estimated to be 1.4–2.2 times more transmissible and able to evade 25-61% of protective immunity elicited by previous infection with non-P.1 lineages (Faria 2021). These data need to be confirmed.
It is currently not known how many of the second wave cases were the result of reinfections in people who had already been infected with the historical virus during the first wave. Find more information about P.1 on page 38.
While the epidemiological situation in South Africa (B.1351) is currently under control, in Brazil it is not (Figure 9). The P.1 variant (Faria 2021, Naveca 2021) which arose first in November created a huge spike of coronavirus cases in the Northern Brazilian state of Amazonas around Manaus, replaced the historical lineage in less than two months and is now spreading around the country.
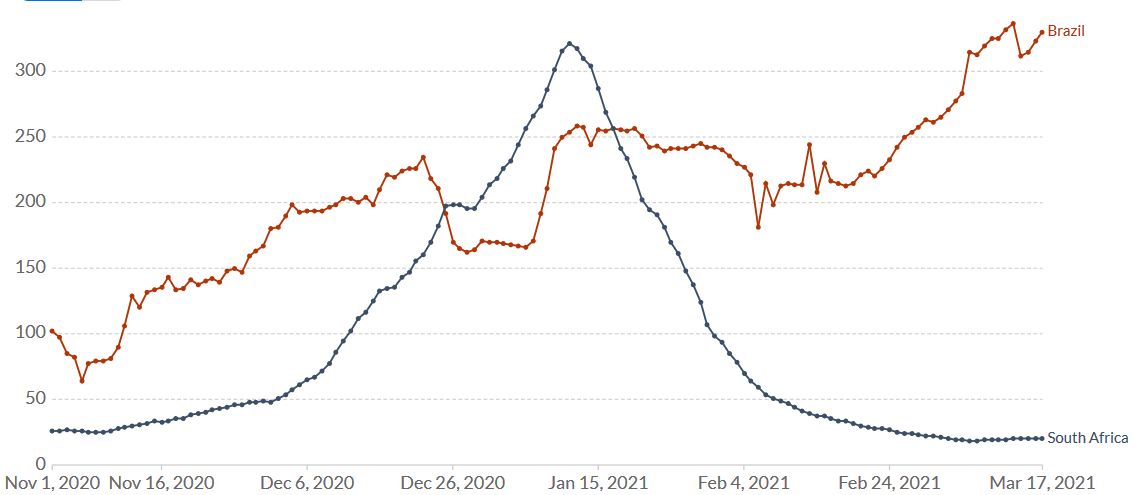
Figure 9. Increase of daily new confirmed COVID-19 cases over the last two weeks in Brazil and South Africa. Shown is the rolling 7-day average per million people. To recalculate these values as the ‘cumulative 7-day rolling incidence per 100.000 people’, multiply the values by 0.7. Example: the March 17 value for Brazil, 330, is equivalent to a cumulative 7-day incidence of 231/100.000 people. Source and copyright: Our World in Data, accessed 18 March.
Israel
Israel is giving the world a glimpse into the SARS-CoV-2 vaccine and post-vaccine world. Already four weeks after the peak in mid-January, there was a 55% reduction in cases, 40% reduction in hospitalizations, 35% reduction in critically ill patients, and a 35% reduction in mortality in people 60 years and older were prioritized to get the vaccine first (Segal 2021). In early March, as more than 90% of people aged 50 or older had received the first vaccine dose, they represented only 11% of currently diagnosed new SARS-CoV-2 cases. In contrast, adolescents and children 19 years or younger represented 47% of new cases (Figure 10). In mid-March, 50% of the population was fully vaccinated and an additional 10% had received the first vaccine dose.
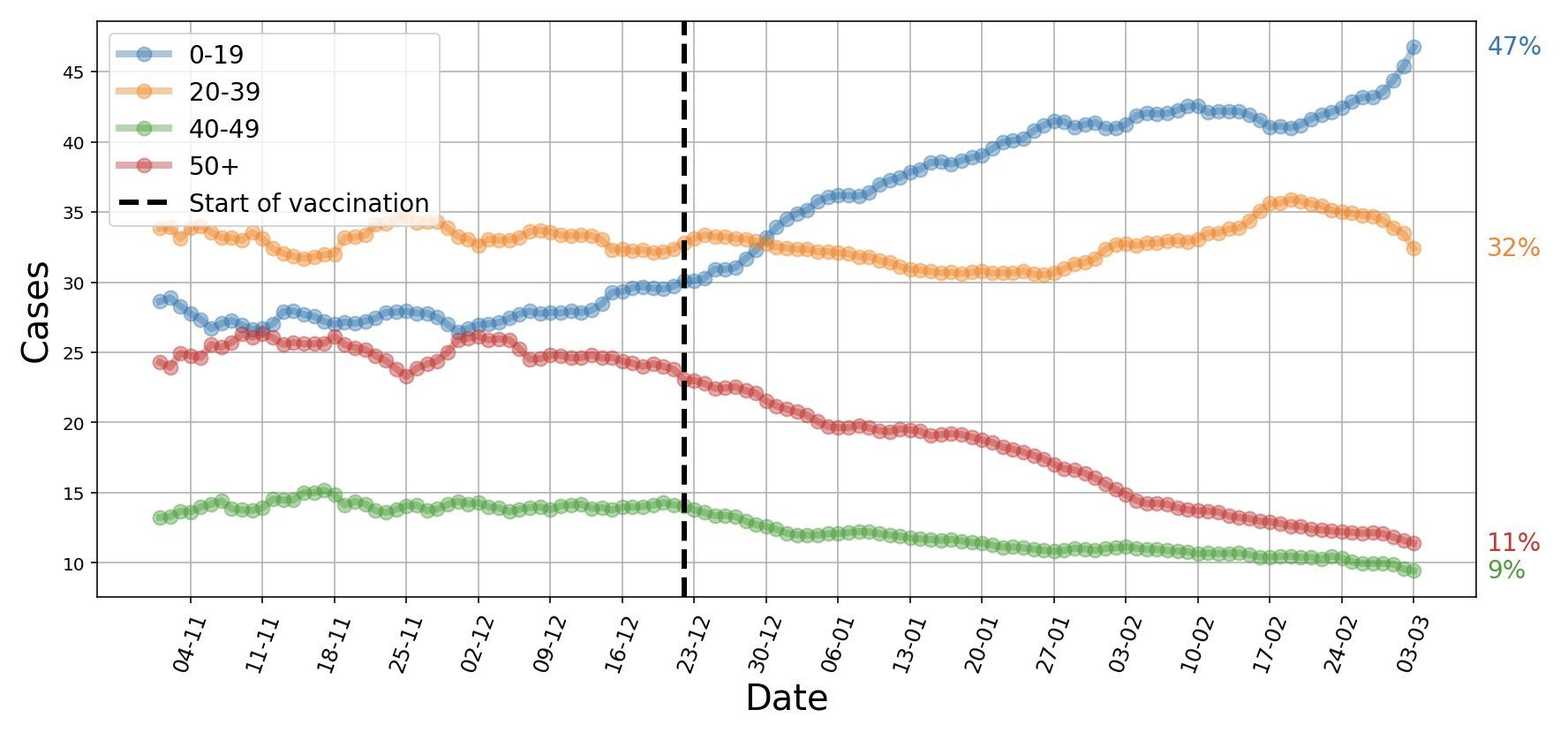
Figure 10. Israel, early March 2021. People 50 years and older represent 11% of currently diagnosed SARS-CoV-2 cases, while adolescents and children 19 years or younger represent 47% of new cases. COVID-19 cases are now younger. Source and copyright: Eran Segal, 5 March, https://bit.ly/3bi6YMF.
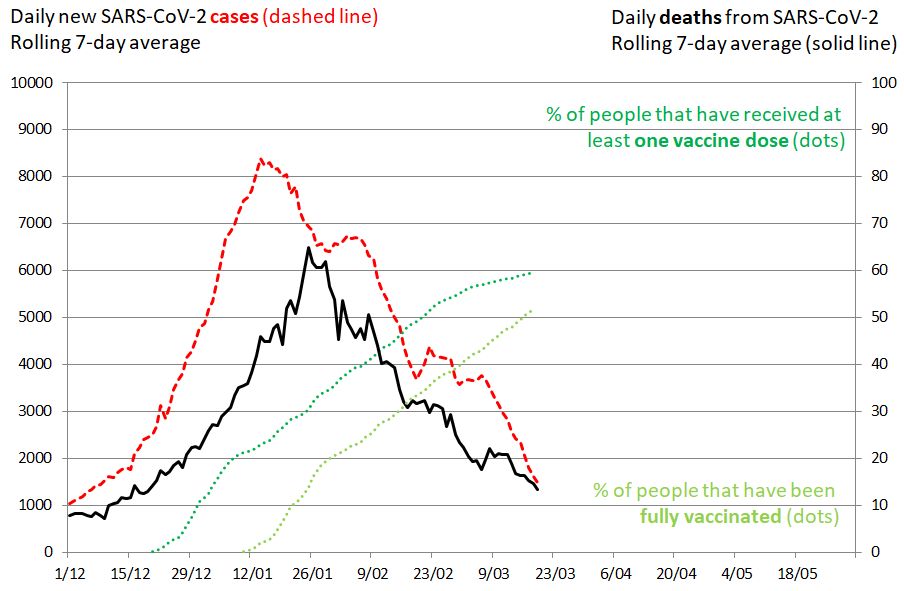
Figure 11. SARS-CoV-2 cases in Israel. 19 March 2021. Impact of mass vaccination on the pandemic. The rolling 7-day average of new SARS-CoV-2 cases is shown in red (left vertical axis), the rolling 7-day average of deaths as a plain black line (right vertical axis). The percentage of people that have received at least one vaccine dose is shown in green. The percentage of people that have been fully vaccinated is shown in green-yellow. The evolution of daily new cases and deaths were influenced by lockdown measures, transmissibility of circulating viruses and the vaccination campaign.
Israel is also an example of how challenging B.1.1.7 can be. Even with an extended vaccination program plus a national lockdown, the third COVID-19 wave is only slowly coming under control. B.1.1.7 has caused the epidemic to drag on for much longer than would have been the case with the historical virus and was partly responsible for a late decline in the number of new cases (Figure 11, red line) and deaths (black line) after the 27 December lockdown. Of note, with more than 2 million children who cannot yet receive the coronavirus vaccine, Israel will not reach herd immunity in the foreseeable future.
US
New York was likely one of the key hubs for introduction and domestic spread of B.1.1.7. (Alpert 2021). The data highlight the relative ease with which SARS-CoV-2 variants can spread undetected throughout the US.
An analysis of 212 B.1.1.7 genomes collected in the US from December 2020 to January 2021 found a doubling rate of a little over a week and an increased transmission rate of 35-45% (Washington 2021) and predict that the US is on a similar trajectory as other countries where B.1.1.7 rapidly became the dominant SARS-CoV-2 variant. Updated data are available at Helix 202102 (Figure 18).
In the US, as in other countries, a quick roll-out of vaccines and continuing non-pharmaceutical interventions such as mask mandates and bans on gatherings will be needed to mitigate the impact of the emerging variants (Bosetti 2021). Due to massive vaccination, the number of hospitalizations will be reduced, but this positive effect of the vaccines may not be sufficient to compensate for the deleterious impact of the variants in the coming two to three months. Some US states will manage to keep the coming B.1.1.7 at bay, but others like Florida will probably not. A weekly updated COVID tracker is available at the CDC website (CDC Tracker 2021).
Transmission
Compared to historical lineages, all variants have a competitive advantage, either because they are more transmissible or because they escape the immune response. The impact on transmission may differ from variant to variant, though. While the transmission advantage of B.1.1.7 and P.1 seems to be 50% or higher (Davies 2021, Volz 2021, Leung 2021, Public Health England 20210126, Gaymard 2021, Faria 2021), the transmission dynamics for B.1.351 are not yet fully understood.
Increased transmission in high-risk groups leads to more SARS-CoV-2 infections, hospitalizations and deaths. Figure 12 depicts a simplified scenario showing the number of new deaths every six days from three different viral strains, assuming each strain started from 10.000 infections. It shows how a more infectious virus may lead to more deaths.
There is evidence that vaccines may reduce transmissibility of SARS-CoV-2. Two studies show a decrease of viral load by 1.6x to 20x in individuals who were positive for SARS-CoV-2 (Petter 2021, Levine-Tiefenbrun 2021).
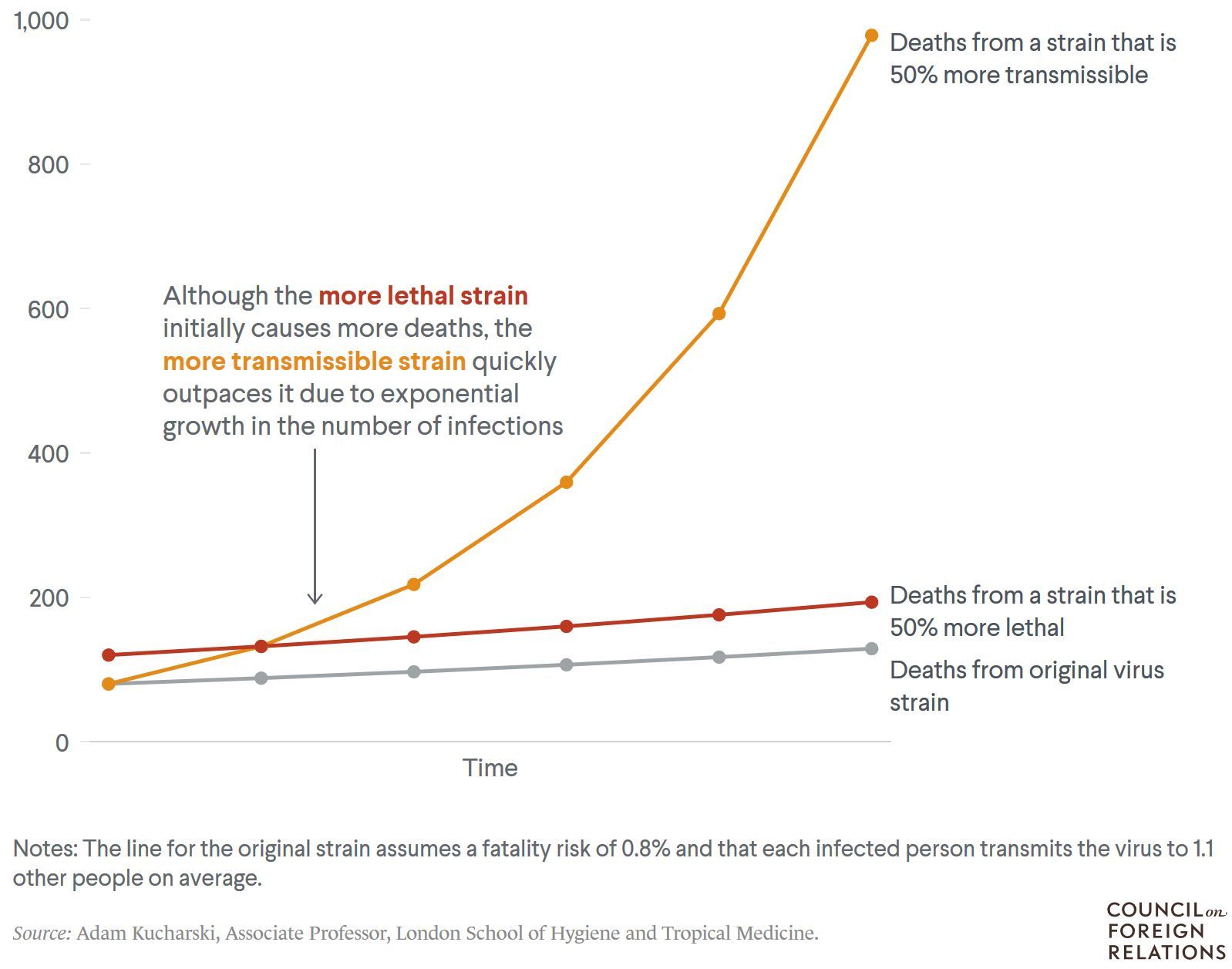
Figure 12. A more infectious virus could lead to many more deaths. Simplified, hypothetical scenario showing the number of new deaths every six days from three different virus strains, assuming each strain started from 10.000 infections. Source: Adam Kucharski, Associate Professor, London School of Hygiene and Tropical Medicine.
Clinical Consequences
Infection with B.1.1.7 is likely to be associated with an increased risk of hospitalization and death compared to infection with historical lineages (NERVTAG 20210211).
In the first study, for a male aged 55–69, the absolute risk of death would increase from 0,6% to 0,9% over the 28 days following a positive test in the community (Davies 2021).
Another study analyzed 184,786 B.1.1.7 positive people from a research platform that covers 40% of England’s population registered with a general practitioner. After controlling for co-morbidities, age, week, region & other sociodemographics, the authors found a 67% increased risk of death for B.1.1.7 compared with non-B.1.1.7 cases (Grint 2021).
Still another study matched 54,906 pairs of participants who tested positive for SARS-CoV-2 between 1 October 2020 and 29 January 2021, followed up until 12 February 2021. The mortality hazard ratio associated with infection with B.1.1.7 compared with infection with historical variants was 1.64 in patients who tested positive for COVID-19 in the community (Challen 2021).
There are as yet no data on different clinical outcomes after infection with B.1.351 or P.1.
Over the coming months, mass vaccination of older age classes will change the profile of the epidemic as proportionally more young people will need hospitalization and ICU care. Individuals less than 60 or even less than 50 years old might account for the majority of hospitalizations and intensive care. Table 1 showing the percentage of hospitalized patients and those requiring ICU treatment in France on 10 March 2021 will serve as a reference for changes to come.
| Table 1. Percentage of hospitalized patients and patients requiring ICU treatment | ||||||||
| Age group | 20-29 | 30-39 | 40-49 | 50-59 | 60-69 | 70-79 | 80-89 | 90+ |
| Hospitalization | 1 | 2 | 4 | 9 | 17 | 24 | 29 | 13 |
| Intensive care | 1 | 2.5 | 6 | 17 | 32 | 32 | 8 | 1 |
Immune Escape
The Question of the Year 2021 is, “Will immunity – natural or vaccine-induced – be effective against evolving SARS-CoV-2 variants or will the variants be able to escape human immunity?” In other words: will B.1.1.7, B.1.351, P.1 or other upcoming variants be able to
- Escape naturally acquired immunity
- Escape vaccine induced immunity
- Escape the effect of monoclonal antibodies
Escape from acquired immunity
There is growing evidence for a long-lived and robust T cell immunity generated after natural SARS-CoV-2 infection (Neidleman 2020). Re-infections with phylogenetically distinct SARS-CoV-2 strains that have been reported were usually milder than the first episode (To 2020, Gupta 2020, Van Elslande 2020, Tillett 2021, Iwasak 2021), with only occasionally more severe disease being reported (Larson 2020). These reports are certainly only the tip of an iceberg of hundreds or thousands of potentially undetected re-infections worldwide; however, today, one year after the first COVID-19 wave, there is no documented epidemic of re-infections in countries where B.1.1.7 has largely replaced the previously circulating SARS-CoV-2 strains. In countries where B.1.351 (South Africa) or P.1 (Brazil) are prevalent, hard epidemiological data that would prove that re-infection with newly emerging variants is a widespread phenomenon do not exist.
In the coming months it will be crucial to understand 1) if variants can partially evade antibodies generated in response to vaccination or previous infection and 2) the extent to which they might contribute to the transmission of SARS-CoV-2 in previously exposed populations. While in vitro studies showed that convalescent plasma samples from 20 patients maintained activity against B.1.1.7 while losing activity against B.1.351 (Wang P 2021), the real-world outcome will largely depend on cellular immune responses. A study by Dan et al. suggested that after SARS-CoV-2 infection (or after vaccination), the vast majority of people could be protected from severe COVID-19 for years and to a certain degree also against variants. An analysis of multiple compartments of circulating immune memory to SARS-CoV-2 in 188 COVID-19 cases showed that spike-specific memory B cells were more abundant at 6 months than at 1 month post symptom onset and SARS-CoV-2-specific CD4+ T cells and CD8+ T cells declined with a half-life of 3-5 months (Dan 2021). Even when antibody neutralization is reduced (and most evident in B.1.351), T cell response could be directed against epitopes conserved across several strains (Skelly 2021; but see also Agerer 2021). Another pre-print study suggests that that CD4+ and CD8+ T cell responses in convalescent COVID-19 subjects or COVID-19 mRNA vaccinees are not substantially affected by mutations found in the SARS-CoV-2 variants (Tarke 2021). A loss of neutralising antibodies (NAb) in plasma may also be countered by the maintenance of neutralising capacity in the memory B cell repertoire (Abayasingam 2021).
For a discussion about innate (Schultze & Aschenbrenner 2021) and adaptive immune responses (Sette & Crotty 2021), see these two reviews.
Escape from vaccine-induced immunity
B.1.1.7
The good news: B.1.1.7, the variant first detected in England, does not escape vaccine-induced immunity (or only unsignificantly so). Real-life data from Israel demonstrate that the Pfizer-BioNTech vaccine was highly effective even in a situation where B.1.1.7 was the dominant lineage. In a trial involving 596,618 vaccinees, the estimated effectiveness 7 days after the second dose was 92% for documented infection, 94% for symptomatic COVID-19, 87% for hospitalization, and 92% for severe COVID-19. Vaccine effectiveness started even two weeks after the first dose (Table 2).
| Table 2. Estimated vaccine effectiveness against COVID-19 outcomes a) 14 to 20 days after the first dose; b) 21 to 27 days after the first dose; and c) 7 days after the second dose. Adapted from Dagan N, Barda N, Kepten E, et al. BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. NEJM February 24, 2021. https://www.nejm.org/doi/full/10.1056/NEJMoa2101765 | |||||
| Period | Documented Infection | Symptomatic COVID-19 | Hospitalization | Severe Disease | Death |
| 14 to 20 days after first dose | 46% | 57% | 74% | 62% | 72% |
| 21 to 27 days after first dose | 60% | 66% | 78% | 80% | 84% |
| 7 days after the second dose | 92% | 94% | 87% | 92% | NA |
How these raw figures translate into day-to-day graphics, is shown by Eran Segal, Hagai Rossman and colleagues who documented a 41% drop in COVID-19 infections in people aged 60 or older from mid-January to early February. During the same period, there was also a 31% drop in hospitalizations (Rossmann 2021, Figure 20). In people aged 59 and younger who received the vaccine later, cases dropped by only 12% and hospitalizations by 5%. The share of people aged 60 or older among those hospitalized for COVID-19 has been constantly falling since 15 January (Figure 14).
Figure 13. Comparison between the population aged 0-59 years old (orange line in A-D) and the population aged > 60 years old (blue line in A-D) during the vaccination period, on a nationwide level. Note: Figures A-D are presented with 2 different y-axis scales. A. Rolling weekly sum of new positive cases. B. Rolling weekly sum of new moderate or severe hospitalizations. C. Rolling weekly sum of new mild, moderate or severe hospitalizations. D. Rolling weekly sum of new severe hospitalizations. Source and copyright: Eran Segal, Hagai Rossmann, et al. – Link: http://bit.ly/36KhjOU
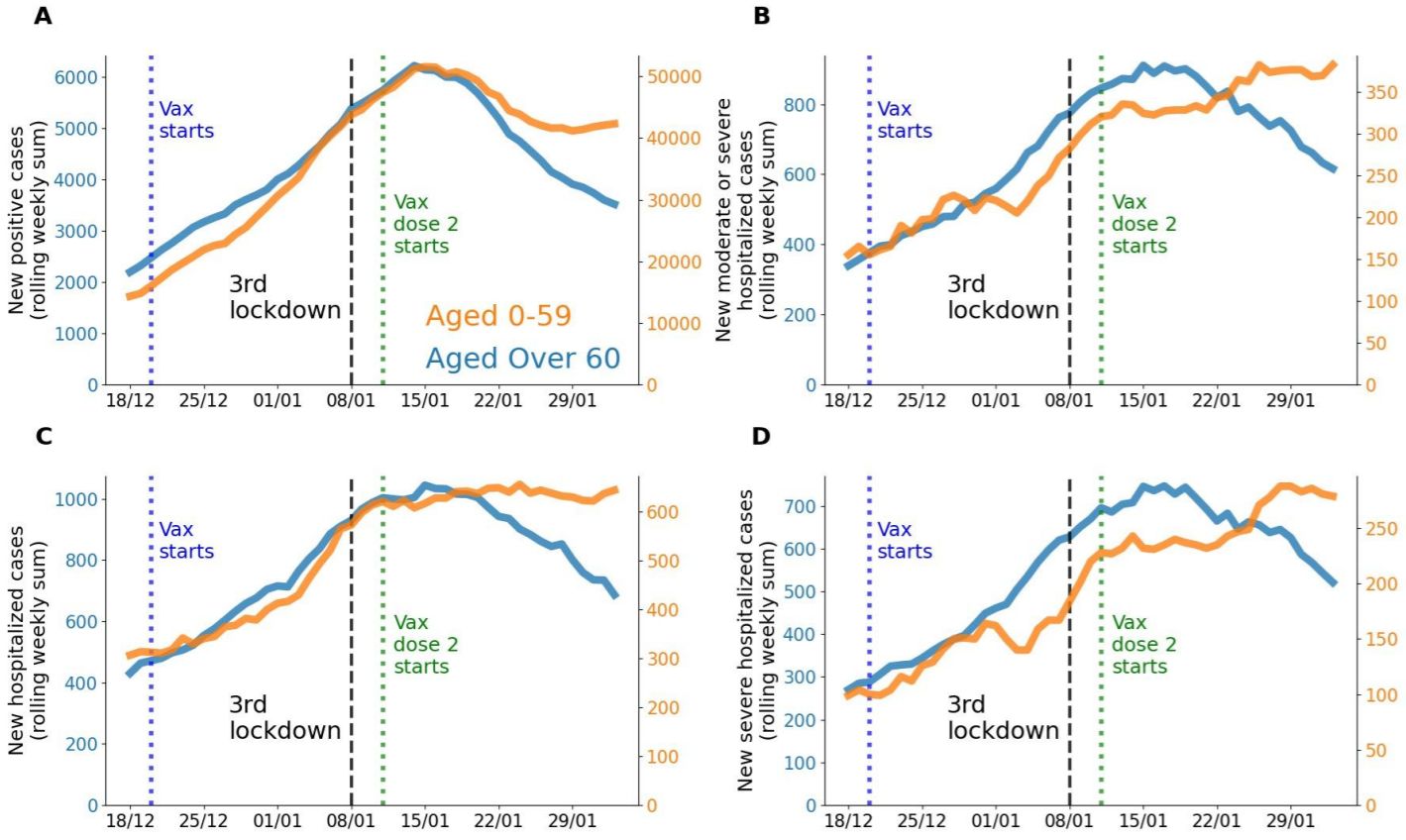
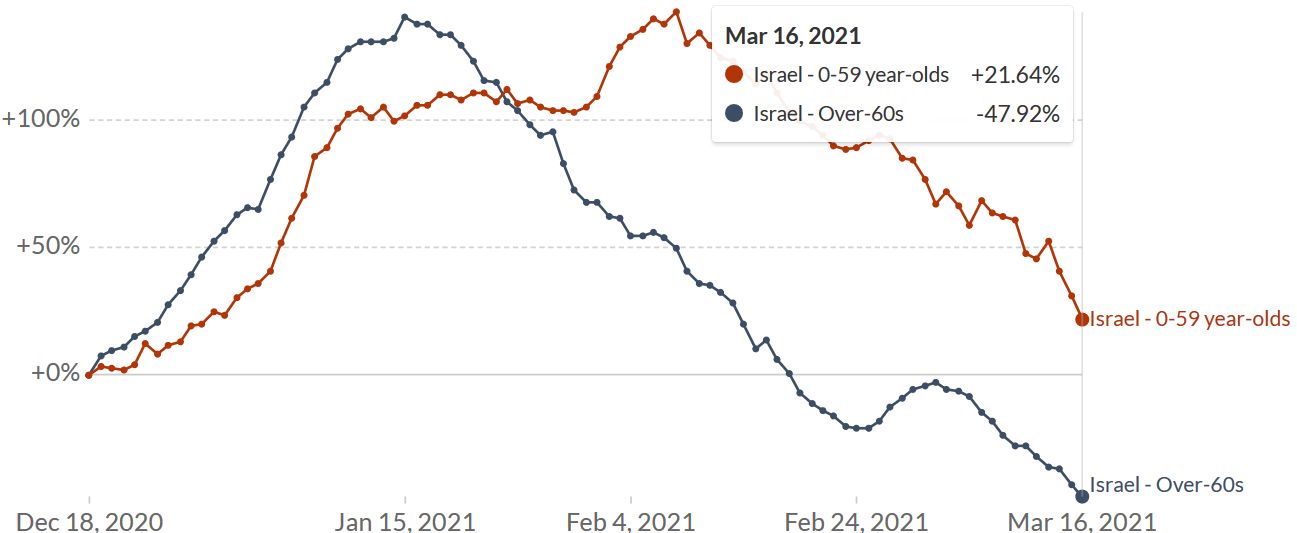
Figure 14. Israel – New hospitalizations for COVID-19 by age. Shown is the rolling weekly sum of COVID-19 hospitalizations. Data is available at the national level, plus breakdown by regions where vaccination began early or late. Source: Our World in Data, accessed 18 March. Link: https://ourworldindata.org/vaccination-israel-impact
Data coming in from Scotland and England confirm these results. The vaccines used there – Pfizer-BioNTech and AstraZeneca – protected well over 80% of vaccinees against COVID-19 related hospitalisation at 28-34 days post-vaccination, even aged ≥ 80 years (Vasileiou 2021, Hall 2021, Public Health England 20210222), and even after a single dose. The Pfizer-BioNTech vaccine might also be effective at preventing infection (66%-85% according to a preprint; Aran 2021) and thus contribute to reduce transmission of SARS-CoV-2.
B.1.351-like variants
B.1.351-like variants include currently B.1.351 (first detected in South Africa) and P.1 (Brazil). Both strains harbor the E484K mutation (Tegally 2021, Voloch 2020). A map of all amino acid mutations to the SARS-CoV-2 spike receptor-binding domain (RBD) showed that the site where mutations tend to have the largest effect on antibody-binding and neutralization is E484 (Greaney 2021b).
| Table 3. Vaccine efficacy against new variants. Adapted from Eric Topol, https://bit.ly/3d3ZmPj, 7 February. | ||
| Vaccine manufacturer | Participants | Main efficacy findings |
| Efficacy against B.1.1.7 | ||
| Novavax | 15,203 | 86% efficacy (vs 96% for historical variant) |
| AstraZeneca | 4236 | 75% efficacy (vs 85% for historical variant) |
| Efficacy against B.1.351 | ||
| Janssen (Johnson & Johnson) |
~10,900 | 57% efficacy (72% in US) |
| Novavax | 4422 | 49% efficacy
– HIV negative: 55% – HIV positive: probably substantially lower |
| AstraZeneca | ~2000 | “minimal protection vs mild-moderate infection” |
There are no population-wide vaccine efficacy data for B.1.351-like variants, but it seems increasingly likely that they have a higher potential for evasion of natural or vaccine-induced immunity than B.1.1.7. Results from two clinical vaccine trials – Janssen’s ENSEMBLE and Novavax’s NVX-CoV2373 – have shown that the level of protection against moderate to severe COVID-19 infection was lower in South Africa where B.1.351 has been the predominant variant of late. The Janssen vaccine candidate provided a level of protection against moderate to severe COVID-19 infection of 57% in South Africa and 72% in the United States (JNJ 20210129) (Table 3). The Novavax product provided a level of protection against mild and moderate-to-severe COVID-19 infection of only 49% in South Africa (Novavax 20210311). [The Novavax trial also found that their vaccine candidate was more efficient against the original COVID-19 strain (96.4%) than against B.1.1.7 (86.3%).] In comparison, the results of the AstraZeneca vaccine in South Africa were disappointing – the ChAdOx vaccine did not show protection against mild-moderate COVID-19 due to B.1.351 (Madhi 2021).
In vivo data
For B.1.1.7, the real-life data from the Israeli vaccination campaign were somewhat anticipated by in vitro studies. An analysis of immune sera from individuals vaccinated with the Pfizer-BioNTech vaccine showed that B.1.1.7 seemed unlikely to escape vaccine-mediated protection (Muik 2021). The authors had investigated SARS-CoV-2-S pseudoviruses bearing either the Wuhan reference strain or the B.1.1.7 lineage spike protein with sera of 16 participants in a previously reported trial with the Pfizer-BioNTech vaccine. The immune sera had equivalent neutralizing titers to both variants. These data have been confirmed in a study describing 50% plaque reduction neutralization testing (PRNT50) with samples obtained from 15 participants 2 or 4 weeks after the administration of the second dose of the Pfizer-BioNTech vaccine (Liu Y 2021).
Generally, in vitro data for B.1.351 and P.1 seem to be of greater concern. Both variants harbor the E484K mutation (Tegally 2021, Voloch 2020) which increasingly seems to be the “bad boy on the block”. A map of all amino acid mutations to the SARS-CoV-2 spike receptor-binding domain (RBD) showed that the site where mutations tended to have the largest effect on antibody-binding and neutralization was E484 (Greaney 2021b). In another study by David H. Ho and colleagues found that the serum of 12 people vaccinated with Moderna’s vaccine and 10 people vaccinated with the Pfizer-BioNTech vaccine was six to nine times less potent against B.1.351. Serum from 20 people previously infected with SARS-CoV-2 was 11 to 33 times less potent (Wang P 2021). E484K accounted for much of the effect.
P.1 is refractory to multiple neutralizing monoclonal antibodies and more resistant to neutralization by (first-wave) convalescent plasma (de Souza 2021, Wang P 2021). Five months after booster immunization with the Chinese CoronaVac vaccine, plasma from vaccinated individuals failed to efficiently neutralize P.1 lineage isolates (de Souza 2021). In a pre-print paper, P.1 has been estimated to be 1.4–2.2 times more transmissible and able to evade 25-61% of protective immunity elicited by previous infection with non-P.1 lineages (Faria 2021). These data need to be confirmed.
The current – preliminary – state-of-knowledge has been summarized in the Figure of the Week twittered by Shane Crotty (Figure 15; if you don’t follow Shane, start following him now: https://twitter.com/profshanecrotty). While natural and vaccine-induced immunity should protect against infection with B.1.1.7, but may be insufficient to protect against B.1.351 and P.1. However, even in the absence of antibody neutralization, we could expect some T cell protection (Tarke 2021). And if circulating memory T cells may not be effective in preventing Sars-CoV-2 infection, it can be expected that they reduce COVID-19 severity. In summary (adapted from Shane Crotty, https://bit.ly/3uUXmPX):
- Several vaccines may provide satisfying immunity against these variants
- Most of the vaccines will probably provide excellent protection against hospitalizations/deaths from these variants
- A booster vaccine against these variants is likely to be highly effective
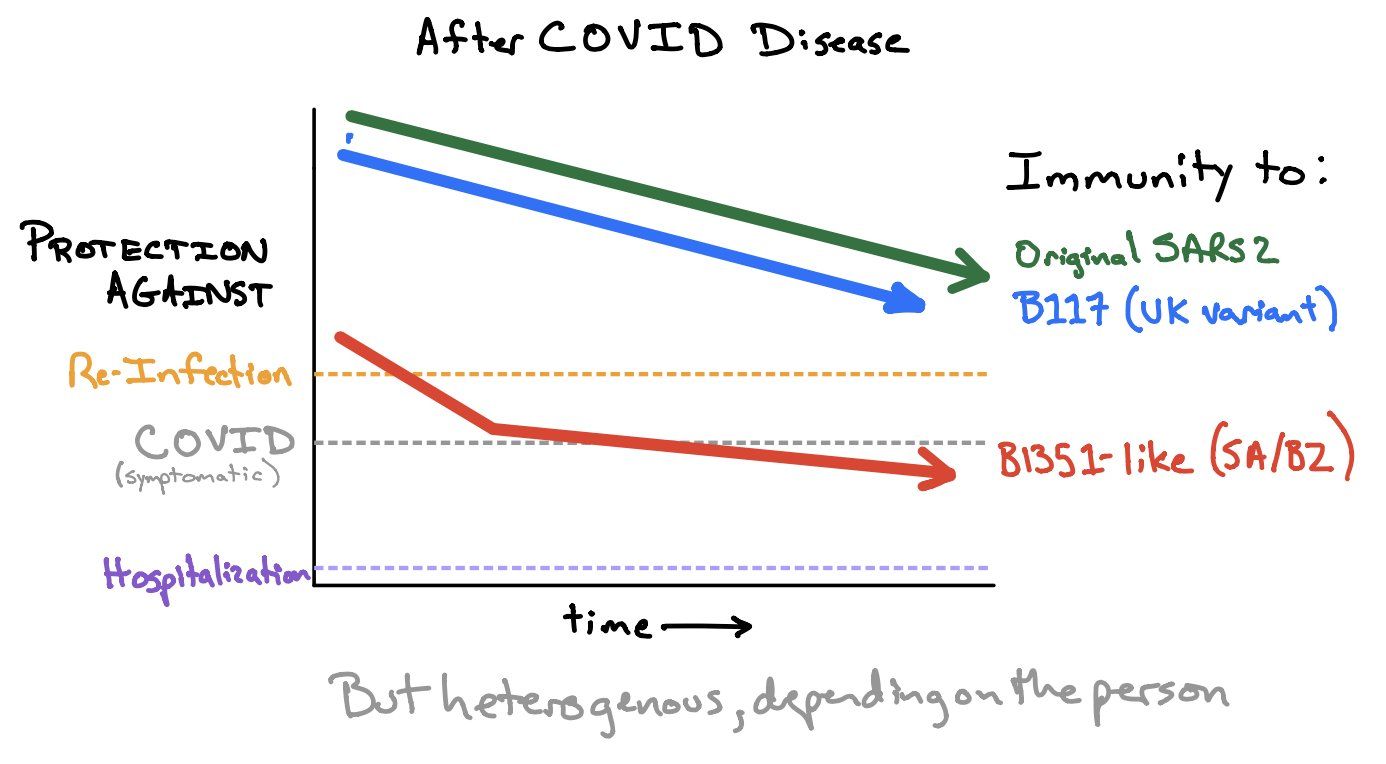
Figure 15. How much immunity do we need to protect against (asymptomatic) re-infection, symptomatic COVID and hospitalization. Source and copyright: Shane Crotty, https://bit.ly/3uUXmPX, March 4.
It is quite probable that SARS-CoV-2 vaccines will need to be reformulated – a challenge most companies have already accepted (see Outlook, page 43). Find more information about immune escape on page 36 (B.1.1.7) and page 37 (B.1.351). There is one piece of good news, though: sera from B.1.351-infected patients have been shown to maintain good cross-reactivity against historical strains (viruses from the first wave) (Cele 2021). Sera from B.1.351-infected patients would also seem to neutralize P.1 (Moyo-Gwete 2021). If these data are confirmed, vaccine manufacturers working on a B.1.351-adapted vaccine are on the right track.
Escape from monoclonal antibodies
Some monoclonal antibodies (mAb) against SARS-CoV-2 infection have received emergency use authorization (Chen 2021, Baum 2020, Hansen 2020, Gottlieb 2021). In November 2020, the FDA issued emergency use authorizations (EUA) for the combination casirivimab plus imdevimab (REGN-CoV2; Regeneron – FDA) and for bamlanivimab (Lilly – FDA) for the treatment of mild-to-moderate COVID-19 and who are at high risk for progressing to severe COVID-19. Other nAbs are still in the pipeline (Ju B 2020, Pinto 2020, Shi R 2020, Zost 2020, Dong J 2021). All face an uncertain future in a world of fast-evolving SARS-CoV-2 variants.
A reminder: Epitope mapping had previously shown that antibodies are divided between those directed against the receptor-binding domain (RBD) of the spike protein and those directed against the N-terminal domain (NTD) of spike, indicating that both of these regions at the top of the viral spike are immunogenic (Liu L 2020). RBD is the prime target of the neutralizing response during infection (Rogers 2020, Piccoli 2020, Barnes 2020, Robbiani 2020) and most antibodies target this region (Piccoli 2020, Tzou 2020). NTD is the next most frequent target of investigational neutralizing antibodies.
Many of the B.1.1.7, B.1.351 and P.1 mutations reside in the RBD (also known as the receptor-binding motif—RBM) or in the antigenic supersite in NTD (Cerutti 2021, McCallum 2021). Recent studies have shown that a single amino-acid mutation (E406W) could fully escape the recently approved REGN-COV2, which consists of two antibodies targeting distinct structural epitopes (Starr 2021). Earlier, there was evidence that one of the spike protein mutations, E484K, might affect neutralization by some polyclonal and monoclonal antibodies (Greaney 2021b, Weisblum 2020). B.1.351 in particular may confer neutralization escape from multiple classes of SARS-CoV-2 directed monoclonal antibodies (Wibmer 2021, Liu Y 2021, Wu K 2021, Wang Z 2021).
Recently, David Ho, Pengfei Wang and colleagues presented a detailed picture of mAb-affecting mutation. After creating VSV-based SARS-CoV-2 pseudoviruses that contained each of the individual mutations as well as one with all 8 mutations of B.1.1.7 (UK∆8) and another with all 9 mutations of B.1.351 (SA∆9), they measured their susceptibility to neutralization by 30 mAbs. The results (see the details in Table 4) are sobering (Wang P 2021):
| Table 4. RBD-directed antibodies. Fold-change in IC50 of neutralizing mAbs against UK∆8 and SA∆9* relative to wild type virus. | |||
| UKΔ8 | SAΔ9 | ||
| RBD**-directed mAbs | 2-36 (Liu L 2020) |
||
| COVA-1 (Brouwer 2020, Liu H 2020) |
|||
| 910-30 (Banach 2021) |
-14.0 | < -1000 | |
| 2-15 (Liu L 2020) |
< -1000 | ||
| Bamlanivimab (LY-CoV555) (Chen P 2021) |
< -1000 | ||
| C121 (Robbiani 2020) |
< -1000 | ||
| Casirivimab (REGN10933) (Baum 2020, Hansen 2020, Weinreich 2020) |
-58,8 | ||
| 2-7 (Liu L 2020) |
|||
| Imdevimab (REGN10987) (Baum 2020, Hansen 2020, Weinreich 2020) |
|||
| C135 (Robbiani 2020) |
|||
| S309 (Pinto 2020) |
-3,1 | ||
| NTD**-directed mAbs | 5-24 (Liu L 2020) |
< -1000 | < -1000 |
| 4-8 (Liu L 2020) |
< -1000 | < -1000 | |
| 4A (Chi X 2020) |
< -1000 | < -406,6 | |
| 2-27 (Liu L 2020) |
-121,2
|
< -1000 | |
| 4-19 (Liu L 2020) |
-20,5 | < -1000 | |
| * David Ho, Pengfai Wang and colleagues at Columbia University produced VSV-based SARS-CoV-2 pseudoviruses that contain each of the individual mutations as well as one with all 8 mutations of the B.1.1.7 variant (UK∆8) and another with all 9 mutations of the B.1.351 variant (SA∆9) and measured its susceptibility to neutralization by 30 mAbs (and also 20 convalescent plasma, and 22 vaccinee sera). For neutralization of UK∆8, only the activities of 910-30 and S309 are impaired, albeit modestly. For neutralization of SA∆9, however, the activities of 910-30, 2-15, LY-CoV555 (bamlanivimab), C121, and REGN10933 (casirivimab) are completely or markedly abolished. Other mAbs such as 2-36, COVA1-16 2-7, REGN10987 (imdevimab), C135, and S309 (which are directed to lower aspects of the “inner or outer side”; see details in the article) retained their activities against SAΔ9.
** RBD: Receptor-binding domain; NTD: N-terminal domain |
|||
- B.1.1.7 (RBD) – For neutralization of UK∆8, only the activities of 910-30 and S309 were impaired, albeit modestly. The decreased activity of 910-30 was mediated by N501Y, whereas the slightly impaired activity of S309 was unexplained.
- B.1.351 (RBD) – For neutralization of SA∆9, however, the activities of 910-30, 2-15, LY-CoV555 (bamlanivimab), C121, and REGN10933 (casirivimab) are completely or markedly abolished. Other mAbs such as 2-36, COVA1-16 2-7, REGN10987 (imdevimab), C135, and S309 (which are directed to lower aspects of the “inner or outer side”; see details in the article) retained their activities against SAΔ9.
Against SAΔ9, the complete loss of activity of 2-15, LY-CoV555, and C121 is mediated by E484K; the complete loss for 910-30 is mediated by K417N; and the marked reduction for REGN10933 is mediated by K417N and E484K.
- B.1.1.7 and B.1.351 (NTD) – Both UKΔ8 and SAΔ9 are profoundly resistant to neutralization by several antibodies.
The resistance of UKΔ8 to most NTD mAbs is largely conferred by 144del, whereas the resistance of SAΔ9 is largely conferred by 242-244del and/or R246I.
In other words, Lilly’s bamlanivimab (LY-CoV555), alone or in combination with CB6, was no longer able to neutralize SAΔ9. While REGN10933+REGN10987 and COV2-2196+COV2-2130 are seemingly unaffected, each of these combinations had a component that lost some neutralizing activity. Although S309 and the Brii-196+Brii-198 combination were not significantly impaired, their potencies were noticeably lower (Wang P 2021). In another study, B.1.351 and P.1 were partially (casirivimab, in REGN-COV2, Regeneron) or fully (bamlanivimab, Lilly) resistant to monoclonal antibodies and was less efficiently inhibited by serum/plasma from convalescent individuals or those vaccinated with the Pfizer-BioNTech vaccine (Hoffmann 2021).
In a more recent study, Jesse Bloom, Tyler Starr and colleague mapped all mutations to the SARS-CoV-2 spike receptor binding domain (RBD) that escape binding by LY-CoV555 (bamlanivimab, a monoclonal antibody manufactured by Lilly), and its cocktail combination with LY-CoV016. Individual mutations that escape binding are present in B.1.351 and P.1 (E484K escapes LY-CoV555, K417N/T escape LY-CoV016). Additionally, the L452R mutation in the B.1.429 lineage escapes LY-CoV555 (Starr 2021).
Conclusion: antibody treatment of SARS-CoV-2 infection might need to be modified in areas where B.1.351 and related variants are prevalent. They also highlight the importance of combination antibody therapy in a context of expanding antigenic SARS-CoV-2 diversity.
Stiff winds ahead for manufacturers of monoclonal antibodies.
The Variants
Nomenclature
The pandemic spread of SARS-CoV-2 has resulted in the generation of tens of thousands of viral genome sequences. From the beginning there was a need for a rational and dynamic viral nomenclature that would account for the expanding phylogenetic diversity of SARS-CoV-2. Such a scheme has been proposed by Andrew Rambaut et al. and is now generally accepted (Rambaut 2020b). The new variants first discovered in the UK, South Africa and Brazil are called B1.1.1.7, B.1.351, and P.1, respectively (Table 5). Common mutations are shown in Table 6. A comparison of mutation in B.1.1.7 and B.1.351 is shown in Figure 16.
| Table 5. The currently circulating variants of concern. Adapted from Eric Topol: https://bit.ly/3tE4Jd8, 17 March | |||
| Official name | B.1.1.7 | B.1.351 | P.1 |
| Where first identified | UK | South Africa | Brazil |
| Other names used in the scientific literature | N501Y.V1 20B/501Y.V1 VOC 202012/1 |
N501Y.V2 20H/501Y.V2 VOC 202012/2 |
N501Y.V3 VOC 202012/3 |
| Mutations | 23 | 21 | 17 |
| Spike mutations | 8 | 9 | 10 |
| Key RBD, spike mutations beyond N501Y in all | 69/70 del, P681H, Y144 del, A570D | E484K, K417N, orf1b deletion | E484K, K417T, orf1b deletion |
| Other mutations, including N-terminal | T716I, S982A, D1118H | L18F, D80A, D215G, ∆242-244, R264I, A701V | L18F, T20N, P26S, D138Y, R190S, H655Y |
| Transmissibility ∆ | > 50% increased | Not established | Not established |
| Lethality ∆ | Likely increased: >30% | ? | ? |
| Immune escape | Unclear | Yes | Yes, but less than B.1.1351 |
| Vaccine efficacy | Minimal reduction
~10% point decline in two trials Novavax: 86% AstraZeneca: 75% |
Yes
J&J: 64% Novavax: 49% (probably substantially less in people living with HIV) AstraZeneca: No efficacy against mild infection |
Yes
J&J: 68% |
| Countries reported | 94 | 48 | 26 |
| Table 6. Shared mutations in B.1.1.7, B.1.351 and P.1. Adapted from Andersen KG: https://bit.ly/2NUvnyy | ||
| B.1.1.7 | B.1.351 | P.1 |
| 69-70 del | L18F | L18F |
| Y144 del | D80A | T20N |
| D215G | P26S | |
| R246I | D138Y | |
| R190S | ||
| K417N | K417T | |
| E484K (still rare) | E484K (Eric) | E484K (Eric) |
| N501Y (Nelly) | N501Y (Nelly) | N501Y (Nelly) |
| A570D | ||
| D614G | D614G | D614G |
| P681H | A701V | H665Y |
| T716I | T1027I | |
| S982A | ||
| D1118H | ||
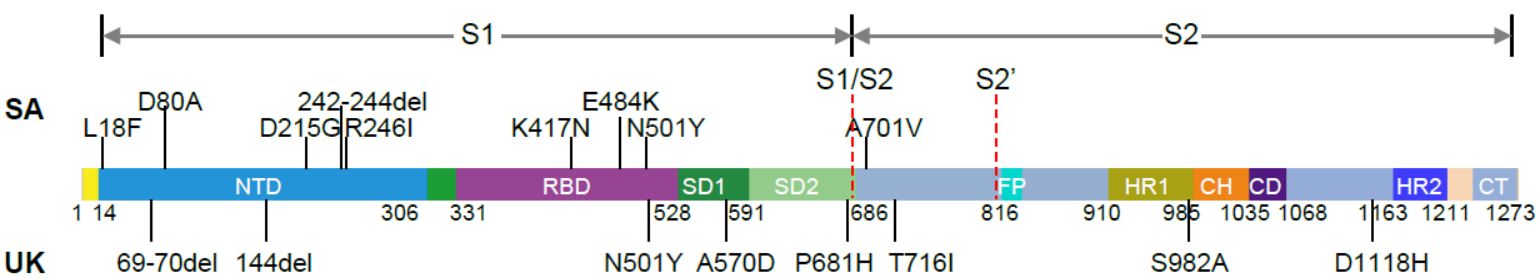
Figure 16. Mutations in the viral spike identified in B.1.351 (SA) and B.1.1.7 (UK) in addition to D614G (Source and copyright: Wang P 2021 – Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. bioRxiv 2021, posted 26 January. Full-text: https://doi.org/10.1101/2021.01.25.428137).
SARS-CoV-2 evolution
Viruses evolve (mutate)
- when they come under pressure, or
- when they are given weeks and months to do so.
Situations of pressure include areas where an explosive epidemic infects large proportions of the population, killing some, but making most people immune. In these settings or in settings where most people, but not all, are immunized through vaccination, only mutant variants that are able to spread despite existing post-infection immunity can sustain continuous chains of transmission.
In an entirely different situation, immunodeficient and chronically infected individuals can be silent incubators of accelerated viral evolution. Such infections are rare, and onward transmission from them presumably even rarer, but they are not improbable (Rambaut 2020, Hensley 2021). High rates of mutations have been reported in immunodeficient or immunosuppressed patients who were chronically infected with SARS-CoV-2. One paper describes the 154-day clinical course of a 45-year-old man with severe antiphospholipid syndrome who was receiving Immunosuppressants medication (Choi 2020). Of interest, amino acid changes were predominantly in the spike gene and the receptor-binding domain, which make up 13% and 2% of the viral genome, respectively, but harbored 57% and 38% of the observed changes. Another report describes the history of an immunocompromised individual with chronic lymphocytic leukemia and acquired hypogammaglobulinemia (Avanzato 2020). In this case, shedding of infectious SARS-CoV-2 was observed up to 70 days.
In the following, we will briefly discuss
- B.1.1.7 (first detected in England)
- B.1.351 (first detected in South Africa)
- P.1 (first detected in Brazil)
B.1.1.7
History and epidemiology
On 14 December 2020, the UK reported to WHO the B.1.1.7 variant (referred to by the UK authorities as SARS-CoV-2 VOC 202012/01: Variant of Concern, year 2020, month 12, variant 01) (WHO 20201231). This variant was first detected in September 2020. Phylogenetic studies carried out by the UK COVID-19 Genomics Consortium soon showed that the new variant had an unusual accumulation of substitutions and was growing at a large rate relative to other circulating lineages (Volz 2021). Within weeks, B.1.1.7 began to replace other viral lineages and as early as November/December 2020, it became the dominant strain in England. As of 20 December 2020, the regions in England with the largest numbers of confirmed cases of the variant were London, the South East, and the East of England (Volz 2021). From there, B.1.1.7 quickly spread all over the country & around the world (Du Z 2021). Between 30 November 2020 and 20 December 2020, 41% of 9321 UK cases that had genomic sequencing data included were B.1.1.7 (Public Health England 20210105). As of 4 March, B.1.1.7 had been identified in 94 countries (Figure 17). A short graphical guide to B.1.1.7 has been published in the lay press by Corum & Zimmer.
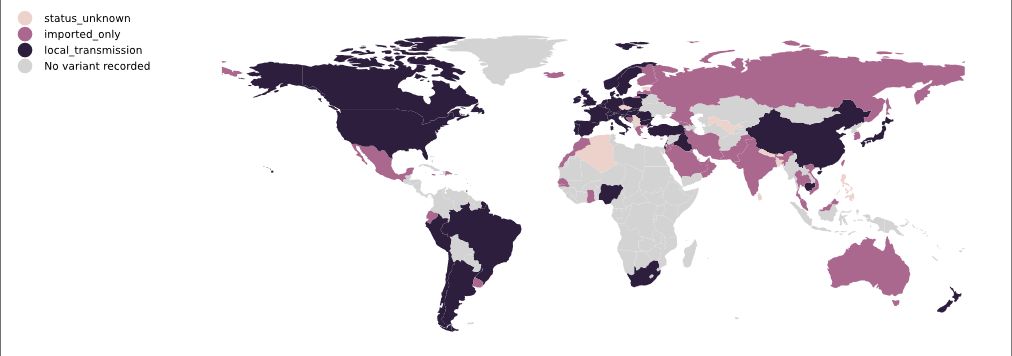
Figure 17. Map of B.1.1.7 local transmission, 20 March 2021 | Colours indicate reports of imported cases (pink) or of local transmission (black). Data is obtained from news reports and similar sources and is manually maintained. Source and copyright: PANGO lineages: B.1.1.7, Áine O’Toole and Verity Hill, Rambaut Group, University of Edinburgh.
Virology
B.1.1.7 has emerged with an unusually large number of mutations in the spike protein (deletion 69-70, deletion 144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H) as well as in other genomic regions (Rambaut 2020). At the moment of discovery, the accrual of 14 lineage-specific amino acid replacements was unprecedented. Three of these mutations are of particular concern:
- Mutation N501Y is one of the six key amino acids interacting with ACE2 receptor and experimental data suggests that it increases ACE2 receptor affinity (Starr 2020). The tyrosine substitution has been shown to have increased binding affinity to the ACE2 receptor (Chan 2020). N501Y has been associated with increased infectivity and virulence in a mouse model (Gu H 2020). [Remember: the receptor binding domain (RBD) of the SARS-CoV-2 spike protein mediates viral attachment to ACE2 receptors. It is a major determinant of host range and a dominant target of neutralizing antibodies.]
- Deletion 69-70 is one of a number of deletions observed in the N terminal domain of the spike protein (McCarthy 2020, Kemp 2020) and is associated with reduced sensitivity to neutralization by SARS-CoV-2 human convalescent serum. It also arose in the mink-associated outbreak in Denmark with the background of the Y453F RBD mutation, and in humans in association with the N439K RBD mutation, accounting for its relatively high frequency in the global genome data (~3000 sequences) (Rambaut 2020).
- Mutation P681H is immediately adjacent to the furin cleavage site between S1 and S2 in spike. [The S1/S2 furin cleavage site of SARS-CoV-2 is not found in other human coronaviruses and has been shown to promote entry into respiratory epithelial cells and transmission in animal models (Hoffmann 2020).]
See also https://covariants.org/variants/S.501Y.V1.
Transmission
Preliminary epidemiologic, modelling, phylogenetic and clinical findings suggest that B.1.1.7 is significantly more transmissible (+50% to +75%) than previously circulating variants (Leung 2020, Volz 2021). It has recently been suggested that B.1.1.7 might cause longer infections with similar peak viral concentration compared to non-B.1.1.7 variants. This extended duration would contribute to B.1.1.7 SARS CoV-2’s increased transmissibility (Table 7) (Kissler 2021).
| Table 7. Longitudinal PCR tests performed in a cohort of 65 individuals infected with SARS-CoV-2 undergoing daily surveillance testing, including seven infected with B.1.1.7 | ||
| B.1.1.7 | non-B.1.1.7 | |
| Mean duration of the proliferation phase |
5.3 days
(2.7. 7.8) (90% credible interval) |
2.0 days
(0.7. 3.3) |
| Mean duration of the clearance phase |
8.0 days
(6.1. 9.9) |
6.2 days
(5.1. 7.1) |
| Mean overall duration of infection (proliferation + clearance phase) |
13.3 days
(10.1. 16.5) |
8.2 days
(6.5. 9.7) |
| Peak viral concentration | 19.0 Ct
(15.8. 22.0) |
20.2 Ct
(19.0. 21.4) |
| log10 RNA copies/ml | 8.5
(7.6. 9.4) |
8.2
(7.8. 8.5) |
Clinical consequences
Infection with B.1.1.7 is most probably associated with an increased risk of hospitalization and death compared to infection with historical strains. Find more on page 20, Clinical Consequences.
Immune escape
See page 21, Immune Escape.
B.1.351
History and epidemiology
On 18 December, national authorities in South Africa announced the detection of a new SARS-CoV-2 variant. The earliest detection had been traced back to October 2020 (South African Government 20201218). It emerged in a severely affected metropolitan area, Nelson Mandela Bay, located on the coast of the Eastern Cape Province. B.1.351 rapidly spread and largely replaced other SARS-CoV-2 viruses circulating in the Eastern Cape, Western Cape, and KwaZulu-Natal provinces. Within weeks it became the dominant lineage in the Eastern Cape and Western Cape Provinces (Tegally 2021, WHO 20201231).
As of 4 March, B.1.351 had been identified in 48 countries (Figure 18).

Figure 18. Map of B.1.351 local transmission, 20 March 2021 | Colours indicate reports of imported cases (pink) or of local transmission (black). Data is obtained from news reports and similar sources and is manually maintained. Source and copyright: PANGO lineages: B.1.351, Áine O’Toole and Verity Hill, Rambaut Group, University of Edinburgh.
Virology
The first description of the B.1.351 lineage found 8 mutations within two immunodominant domains of the spike protein: one cluster in the N-terminal domain (NTD) that includes four substitutions and a deletion (L18F, D80A, D215G, Δ242-244, and R246I), and another cluster of substitutions including three at important residues in the receptor-binding domain (K417N, E484K and N501Y) (Tegally 2021). Unlike the B.1.1.7 lineage detected in the UK, B.1.351 does not contain the deletion at 69/70.
See also https://covariants.org/variants/S.501Y.V2.
Transmission
While the full significance of the B.1.351 mutations described above is not yet clear, the genomic and epidemiological data suggest that this lineage may be associated with increased transmissibility (Tegally 2021). A mathematical model has estimated that B.1.351 could be 50% more transmissible than previously circulating variants in South Africa (Pearson 2021).
Clinical consequences
At this stage, there is no clear evidence to suggest that B.1.351 has any impact on disease severity. A more precise picture will evolve over the next few months.
Immune escape
In a brilliant study, Cele et al. compared neutralization of a first wave historial virus versus the B.1.351 variant using plasma collected from adults hospitalized with COVID-19 from two South African infection waves, with the second wave dominated by B.1.351infections. B.1.351 was poorly neutralized by plasma collected from adults infected with historical SARS-CoV-2 strains. In contrast, plasma samples from the second wave which was dominated by B.1.351 showed only a modest decline relative in neutralization of historical strains (Cele 2021). These data provide preliminary evidence that vaccines based on sequences such as B.1.351 could retain activity against other circulating SARS-CoV-2 lineages.
See also page 21, Immune Escape.
P.1
History and epidemiology
On January 6, 2021, the National Institute of Infectious Diseases (NIID) of Japan detected a new variant isolate of SARS-CoV-2 in isolates collected at airport screening from four travelers who arrived in Tokyo from Amazonas, Brazil, on January 2, 2021 at airport screening. The isolate had some mutations found in previously reported variant isolates of concern from the UK and South Africa (Fujino 2021). The new variant isolate had 12 mutations in the spike protein, including N501Y and E484K. A few days later, on 12 January 2021, a pre-print article described a variant detected in Manaus, Brazil, identical to the one detected in Japan (Faria 2021). The new variant, P.1, was identified in 42% (13 out of 31) of RT-PCR positive samples collected between 15 and 23 December in Manaus (Faria 2021). At the time, Manaus was experiencing an upsurge in COVID-19 cases.
As of 4 March, P.1 had been identified in 26 countries (Figure 19).
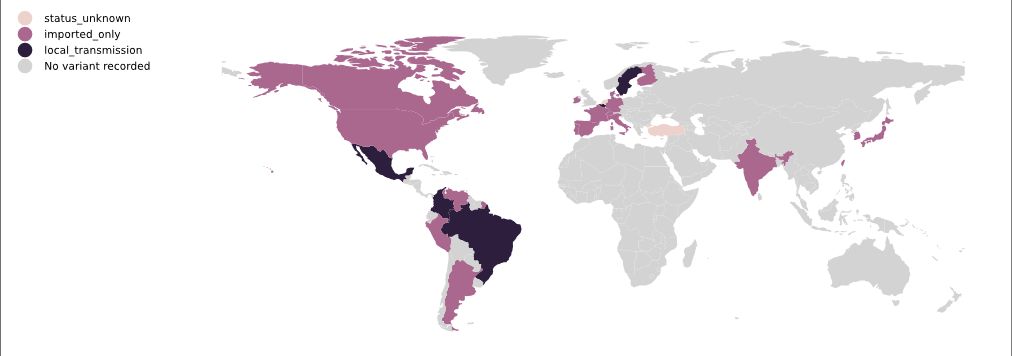
Figure 19. Map of P.1 local transmission, 4 March 2021 | Colours indicate reports of imported cases (pink) or of local transmission (black). Data is obtained from news reports and similar sources and is manually maintained. Source and copyright: PANGO lineages: P.1, Áine O’Toole and Verity Hill, Rambaut Group, University of Edinburgh.
Virology
The new P.1 lineage carries 17 unique amino acid changes, 3 deletions, and 4 synonymous mutations, and one 4nt insertion compared to the most closely related available non-P.1 sequence (EPI_ISL_722052) (Faria 2021). P.1 has 11 amino acid changes in the spike protein compared to its ancestral lineage B.1.1.28 (L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, H655Y, T1027I, and V1176F). Some mutations, such as E484K, N501Y and K417T, might influence antibody and vaccine efficacy. The variant is not closely related to B.1.1.7 or B.1.351.
The P.1 lineage and B.1.1.7 (first described in the UK – Rambaut 2020) share the spike N501Y mutation and a deletion in ORF1b (del11288-11296, 3675-3677 SGF). P.1 and B.1.351 (first described in South Africa – Tegally 2021) share three mutation positions in common in the spike protein (K417N/T, E484K, N501Y) (Faria 2021). Both the P.1 and the B.1.351 lineage also have the orf1b deletion del11288-11296 (3675-3677 SGF) (Faria 2021).
The P.1 variant also is known as B.1.1.28 in the Phylogenetic Assignment of Named Global Outbreak Lineages (https://cov-lineages.org/pangolin.html) and as 20J/501Y.V3 in NextStrain (https://nextstrain.org).
Another variant detected in Rio de Janeiro, Brazil, P2, is currently not considered a variant of concern. However, P2 is being intensely investigated because it has the E484K Spike and has been increasing in numbers since October (Resende 2021, Vasques Nonaka 2021, Fiocruz 2021).
See also https://covariants.org/variants/S.501Y.V3.
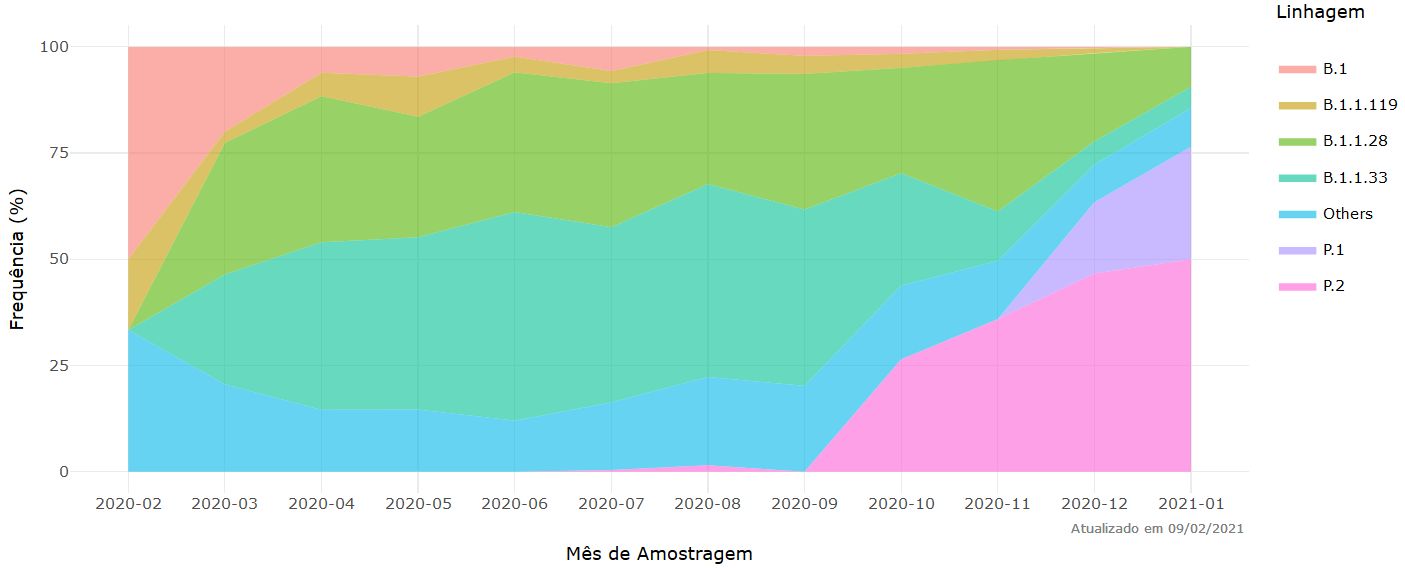
Figure 20. Frequency of the main strains of SARS-Co-2 per month of sampling. Source and copyright: Fiocruz. http://www.genomahcov.fiocruz.br/frequencia-das-principais-linhagens-do-sars-cov-2-por-mes-de-amostragem/
Transmission
Manaus, the largest city in the Amazon region, has seen a dramatic surge in SARS-CoV-2 infettions since mid-December. As this coincides with a report of more than 40% RT-PCR positive P.1 samples collected between 15 and 23 December (Faria 2021), it may be rightly assumed that the new variant has led to an increase in transmissibility of the virus. A previous pre-print paper claiming ‘herd immunity-like’ infection rates in September should be interpreted with caution.
Clinical consequences
At this stage, there is no clear evidence to suggest that P.1 has any impact on disease severity. A more precise picture will evolve within the next months.
Immune escape
See page 21, Immune Escape.
B.1.429 (CAL.20C)
In California, the proportion of SARS-CoV-2 cases associated with this variant rose from 3.8% to 25% between mid-November and late December. By then, B.1.429 (CAL.20C) accounted for 24% of samples in one study, and 36.4% (66/181) of samples in a local Los Angeles cohort (Zhang W 2021). The emerging predominance of this strain is temporally related to the time of onset of the current spike in SARS-CoV-2 infections in Southern California. B.1.429 (CAL.20C) is defined by mutations in the S protein (L452R, S13I, W152C) and in the ORF1a (I4205V) and ORF1b protein (D1183Y).
Mutations
Do you know “what mutations define a variant, what impact they might have (with links to papers and resources), and where variants are found”? If you hesitate a bit, the excellent web site https://covariants.org, by Emma Hodcroft et al, will provide you a primer (Figure 27). See also the JAMA paper by Lauring & Hodcroft, 2021, Genetic Variants of SARS-CoV-2—What Do They Mean? (Laurimg
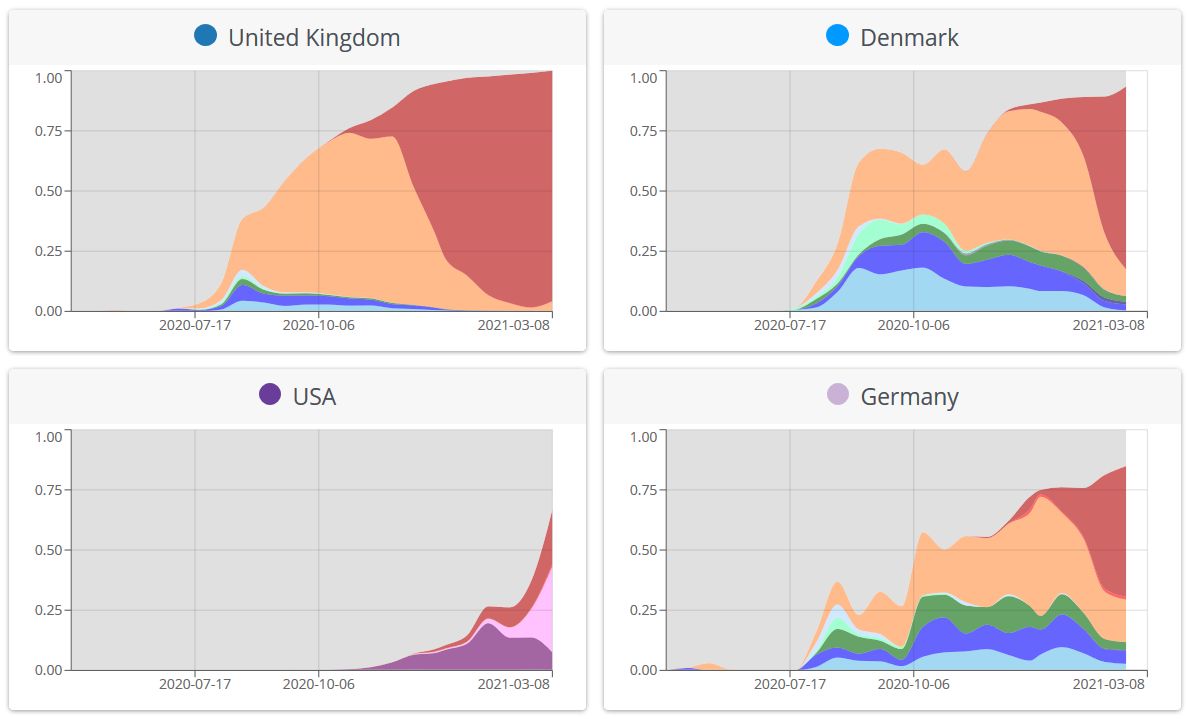
Figure 21. Overview of variants in selected countries. The graphs show for each country the proportion of total number of sequences (not cases), over time, that fall into defined variant groups. Source and copyright: CoVariants.org, by Emma Hodcroft et al.
Prevention and Care
In the coming months, the new SARS-CoV-2 variants will confront many countries with a novel wave of viral spread. Once a more contagious variant has established itself, stabilizing the number of new infections will become increasingly difficult (Priesemann 2021), leading to a spiral of increasing number of infections, hospitalisations and deaths. The increased transmissibility of SARS-CoV-2 variants has far-reaching public health consequences. Non-pharmaceutical interventions (NPIs = everything from mask wearing to lockdowns) which were sufficient to control previous SARS-CoV-2 lineages may need to be reinforced to control B.1.1.7, B.1.351 and P.1. Fortunately, as shown in the Epidemiology section above (page 10), hard lockdowns, including closing of primary schools, secondary schools, and universities (Davies 2021) are effective against new variants. In the future, wastewater analyses may help predict outbreaks with new variants. In Switzerland, a group found evidence for the presence of several mutations that define B.1.1.7 and B.1.351 in a sample from a Swiss ski resort dated around mid-December, two weeks before its first verification in a patient sample in the country (Jahn 2021).
Since the population groups driving transmission will not be targeted with vaccination for some months, ECDC recommends that Member States should to be very cautious about relaxing currently enacted NPIs. Non-essential travel should be avoided. Vaccination should focus on protecting those most at risk from severe disease. Find an overview of Options for response in ECDC’s Rapid Risk Assessment (ECDC 20210121, page 15), in particular
- Surveillance, testing and detection of the emering variants
- Non-pharmaceutical interventions
- Community measures
- Shielding medically and socially vulnerable populations
- Considerations for school settings
- Contact tracing for emerging variants
- Measures for travellers
- Vaccination
- Availability of COVID-19 vaccines
- Monitoring breakthrough infections following vaccination, adjustment of vaccination schedules and possible update of vaccine contents due to SARS-CoV-2 variants in circulation
- Accelerating vaccination campaigns
- Vaccine effectiveness studies
- Hospital and healthcare preparedness
These recommendations translate into 9 commandments: “Whenever possible:
- Stay home, if you can
- Avoid gatherings, both inside and outside your household
- Avoid enclosed spaces
- Wear a mask, do not sing or shout!
- Keep a distance – 2 meters!
- Ventilate whenever you can
- Wash hands
- Disinfect hightouch surfaces (maybe less important?)
- Get vaccinated as soon as you can!”
Or, according to the more joyful words of UN Women: “The pandemic is hard – spread joy:
- Buy someone flowers
- Call a loved one
- Deliver groceries to your neighbour
- Write a greeting card
- Motivate a friend who needs a boost
- Virtually tutor a student
Outlook
In a few months, we will learn more about whether and how B.1.1.7, B.1.351 and P.1 will
- Change the clinical presentation of COVID-19 and mortality
- Affect the few existing treatment option (corticosteroids, tocilizumab, anticoagulants, etc.)
- Increase the number of reinfections
- Affect the immune response to vaccines
- Affect the therapeutical benefit from monoclonal antibodies
Fortunately, global genomic surveillance and rapid open-source sharing of viral genome sequences have facilitated near real-time detection, comparison, and tracking of evolving SARS-CoV-2 variants that can inform public health efforts to control the pandemic (Galloway 2020).
Vaccine producers are at work. Pfizer-BioNTech and Moderna are modifying their vaccine for emerging variant booster candidates. They will test an additional (third) vaccine booster dose to study if neutralizing titers against emerging variants can be increased. All companies have started talks with regulators to know what types of clinical trials and safety reviews would be required to authorize new versions of already approved vaccines. Over time, it is possible that, as with seasonal influenza, these adaptive changes in antigenic regions of the virus would give rise to continual reformulation of existing vaccines (Kistler 2021).
We expected 2021 to the year of the SARS-CoV-2 vaccine. We didn’t expect it to be the year of a race between the virus and SARS-CoV-2 vaccines. Vaccines – and science – will ultimately prevail, but for the coming months, the race will stay close. When the COVID waters calm down in a year or so, we should not stop thinking about infectious diseases. The next pathogen to emerge might be less accommodating (Burton 2021) and we could start thinking about a rational vaccine design based on broadly neutralizing antibodies (Burton & Topol 2021). Creating the tools for preventing the next coronavirus pandemic is within our means and should be considered a global health priority. We can either invest now or pay substantially more later (Koff & Berkley 2021).
References
- Abayasingam A, Balachandran H, Agapiou D, et al. Long-term persistence of RBD-positive memory B cells encoding neutralising antibodies in SARS-CoV-2 infection. Cell Rep Med March 14, 2021. https://www.cell.com/action/showPdf?pii=S2666-3791%2821%2900044-6
- Agerer B, Koblischke M, Gudipati V, et al. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8+ T cell responses. Sci Immunol. 2021 Mar 4;6(57):eabg6461. PubMed: https://pubmed.gov/33664060. Full-text: https://doi.org/10.1126/sciimmunol.abg6461
- Alpert T, Lasek-Nesselquist E, Brito AF, et al. Early introductions and community transmission of SARS-CoV-2 variant B.1.1.7 in the United States. medRxiv 2021, posted 12 February. Full-text: https://doi.org/10.1101/2021.02.10.21251540
- Althaus C, et al. Transmission of SARS-CoV-2 variants in Switzerland. Institute of Social and Preventive Medicine (ISPM), University of Bern 2021, reported 19 February. Full-text: https://ispmbern.github.io/covid-19/variants/
- Amit S, Regev-Yochay G, Afek A, et al. Early rate reductions of SARS-CoV-2 infection and COVID-19 in BNT162b2 vaccine recipients. Lancet February 18, 2021. https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(21)00448-7/fulltext
- Andreano E, Piccini G, Licastro D, et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 Dec 28:2020.12.28.424451. PubMed: https://pubmed.gov/33398278. Full-text: https://doi.org/10.1101/2020.12.28.424451
- Aran D. Estimating real-world COVID-19 vaccine effectiveness in Israel. GitHub 2021, posted 4 February. Full-text: https://github.com/dviraran/covid_analyses/blob/master/Aran_letter.pdf
- Avanzato VA, Matson MJ, Seifert SN, et al. Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell. 2020 Dec 23;183(7):1901-1912.e9. PubMed: https://pubmed.gov/33248470. Full-text: https://doi.org/10.1016/j.cell.2020.10.049
- Bager P, Wohlfahrt J, Fonager J, Albertsen M, et al. Increased Risk of Hospitalisation Associated with Infection with SARS-CoV-2 Lineage B.1.1.7 in Denmark. Lancet Preprints 2021, posted 2 March. Full-text: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3792894
- Bal A, Destras G, Gaymard A, et al. Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France, August to December 2020. bioRxiv 2021, posted 11 January. Full-text: https://doi.org/10.1101/2020.11.10.20228528
- Banach BB, Cerutti G, Fahad AS, et al. Paired heavy and light chain signatures contribute to potent SARS-CoV-2 neutralization in public antibody responses. bioRxiv, 2020.2012.2031.424987 (2021). Full-text: https://doi.org/10.1101/2020.12.31.424987
- Baric RS. Emergence of a Highly Fit SARS-CoV-2 Variant. NEJM December 16, 2020. Full-text: https://doi.org/10.1056/NEJMcibr2032888
- Barnes CO, West AP Jr, Huey-Tubman KE, et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020 Aug 20;182(4):828-842.e16. PubMed: https://pubmed.gov/32645326. Full-text: https://doi.org/10.1016/j.cell.2020.06.025
- Bartsch M, Bohr F, von Bredow R, et al. Can We Stop a Super Coronavirus? Der Spiegel 2021, published 19 January. Full-text: https://www.spiegel.de/international/world/can-germany-stop-the-new-supervirus-a-e9ffc207-0015-4330-8361-b306f6053e15
- Baum A, Fulton BO, Wloga E, et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020 Aug 21;369(6506):1014-1018. PubMed: https://pubmed.gov/32540904. Full-text: https://doi.org/10.1126/science.abd0831
- Beale R. Eeek! LRB 2021, published 4 March. Full-text: https://www.lrb.co.uk/the-paper/v43/n05/rupert-beale/eeek
- Berndt C, Endt C, Müller-Hansen S. Die unsichtbare Welle. Süddeutsche Zeitung 2021, published 5 February. Full-text: https://www.sueddeutsche.de/wissen/coronavirus-mutante-B.1.1.7-daten-1.5197700
- Borges V, Sousa C, Menezes L, et al. Tracking SARS-CoV-2 VOC 202012/01 (lineage B.1.1.7) dissemination in Portugal: insights from nationwide RT-PCR Spike gene drop out data. Virological.org 2021, published 19 January. Full-text: https://virological.org/t/tracking-sars-cov-2-voc-202012-01-lineage-b-1-1-7-dissemination-in-portugal-insights-from-nationwide-rt-pcr-spike-gene-drop-out-data/600
- Borges V, Sousa C, Menezes L, et al. Tracking SARS-CoV-2 VOC 202012/01 (lineage B.1.1.7) dissemination in Portugal: insights from nationwide RT-PCR Spike gene drop out data. Virological.org 2021, published 19 January. Full-text: https://virological.org/t/tracking-sars-cov-2-voc-202012-01-lineage-b-1-1-7-dissemination-in-portugal-insights-from-nationwide-rt-pcr-spike-gene-drop-out-data/600
- Bosetti P, Tran Kiem C, Andronico A, et al. A race between SARS-CoV-2 variants and vaccination: The case of the B.1.1.7 variant in France. Institut Pasteur 2021, posted 23 February. Full-text: https://hal-pasteur.archives-ouvertes.fr/pasteur-03149525
- Brouwer PJM, Caniels TG, van der Straten K, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020 Aug 7;369(6504):643-650. PubMed: https://pubmed.gov/32540902. Full-text: https://doi.org/10.1126/science.abc5902
- Brunning A. How are RNA vaccines made? Periodic Graphics 2021, published 3 January. Link: https://cen.acs.org/pharmaceuticals/vaccines/Periodic-Graphics-RNA-vaccines-made/99/i1
- Burton DR, Topol EJ. Variant-proof vaccines — invest now for the next pandemic. Nature 2021, published 8 February. Full-text: https://www.nature.com/articles/d41586-021-00340-4
- Burton DR, Topol EJ. Variant-proof vaccines — invest now for the next pandemic. Nature 2021, published 8 February. Full-text: https://www.nature.com/articles/d41586-021-00340-4
- Buss LF, Prete Jr A, Abrahim CMM, et al. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science 2020b, published 8 December. Full-text: https://doi.org/10.1126/science.abe9728
- Buss LF, Prete Jr CA, Abrahim CMM, et al. COVID-19 herd immunity in the Brazilian Amazon. medRxiv 2020, posted 21 September. Full-text: https://doi.org/10.1101/2020.09.16.20194787
- Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006 Jul;19(3):531-45. PubMed: https://pubmed.gov/16847084. Full-text: https://doi.org/10.1128/CMR.00017-06
- Callaway E 20210108. Could new COVID variants undermine vaccines? Labs scramble to find out. Nature 2021, published 8 January. Full-text: https://www.nature.com/articles/d41586-021-00031-0
- Callaway E 20210121. Fast-spreading COVID variant can elude immune responses. Nature News 21 January 2021. Full-text: https://www.nature.com/articles/d41586-021-00121-z
- CDC 2021 NCV. New COVID-19 Variants. Centers for Disease Control 2021 – accessed 23 January 2021. Full-text: https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant.html
- CDC 2021 V. Emerging SARS-CoV-2 Variants. Centers for Disease Control 2021 – accessed 23 January 2021. Full-text: https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html
- CDC Tracker. COVID Data Tracker Weekly Review. CDC 2021, updated 19 February, accessed 20 February. Full-text: https://www.cdc.gov/coronavirus/2019-ncov/covid-data/covidview/index.html
- Cele S, Gazy I, Jackson L, et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. MedRxiv 2021, posted 27 February. Full-text: https://doi.org/10.1101/2021.01.26.21250224
- Cerutti G, Guo Y, Zhou T, et al. Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite. bioRxiv 2021, posted 11 January. Full-text: https://doi.org/10.1101/2021.01.10.426120
- Challen R, Brooks-Pollock E, Read JM, Dyson L, Tsaneva-Atanasova K, Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021 Mar 9;372:n579. PubMed: https://pubmed.gov/33687922. Full-text: https://doi.org/10.1136/bmj.n579
- Chan KK, Tan TJC, Narayanan KK, Procko E. An engineered decoy receptor for SARS-CoV-2 broadly binds protein S sequence variants. bioRxiv. 2020 Dec 21:2020.10.18.344622. PubMed: https://pubmed.gov/33398275. Full-text: https://doi.org/10.1101/2020.10.18.344622
- Chandrashekar A, Liu J, Martinot AJ, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020 May 20:eabc4776. PubMed: https://pubmed.gov/32434946. Full-text: https://doi.org/10.1126/science.abc4776
- Chen P, Nirula A, Heller B, et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N Engl J Med. 2021 Jan 21;384(3):229-237. PubMed: https://pubmed.gov/33113295. Full-text: https://doi.org/10.1056/NEJMoa2029849
- Chi X, Yan R, Zhang J, et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020 Aug 7;369(6504):650-655. PubMed: https://pubmed.gov/32571838. Full-text: https://doi.org/10.1126/science.abc6952
- Choi B, Choudhary MC, Regan J, et al. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N Engl J Med. 2020 Dec 3;383(23):2291-2293. PubMed: https://pubmed.gov/33176080. Full-text: https://doi.org/10.1056/NEJMc2031364
- Contreras S, Priesemann V. Risking further COVID-19 waves despite vaccination. Lancet Infect Dis 2021, published 18 March. Full-text: https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(21)00167-5/fulltext
- Corum J, Zimmer C. Inside the B.1.1.7 Coronavirus Variant. The New York Times 2021, published 18 January. Full-text: https://www.nytimes.com/interactive/2021/health/coronavirus-mutations- B.1.1.7-variant.html
- COVID-19 Genomics UK (COG-UK) consortiumcontact@cogconsortium.uk. An integrated national scale SARS-CoV-2 genomic surveillance network. Lancet Microbe. 2020 Jul;1(3):e99-e100. PubMed: https://pubmed.gov/32835336. Full-text: https://doi.org/10.1016/S2666-5247(20)30054-9
- da Silva Francisco R, Benittes F, Lamarca AP, et al. Pervasive transmission of E484K and emergence of VUI-NP.13L with evidence of SARS-CoV-2 co-infection events by two different lineages in Rio Grande do Sul, Brazil. medRxiv 2021, posted 26 January. Full-text: https://doi.org/10.1101/2021.01.21.21249764
- Dagorn G. Covid-19 : comment le variant B.1.1.7, apparu en Angleterre, menace de relancer l’épidémie en France. Le Monde 2021, published 19 February. Full-text : https://www.lemonde.fr/les-decodeurs/article/2021/02/19/comment-le-variant-britannique-menace-de-destabiliser-l-epidemie-en-france_6070563_4355770.html
- Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021 Feb 5;371(6529):eabf4063. PubMed: https://pubmed.gov/33408181. Full-text: https://doi.org/10.1126/science.abf4063
- Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021 Mar 3:eabg3055. PubMed: https://pubmed.gov/33658326. Full-text: https://doi.org/10.1126/science.abg3055
- Davies NG, Jarvis CI, CMMID COVID-19 Working Group, et al. Increased hazard of death in community-tested cases of SARS-CoV-2 Variant of Concern 202012/01. medRxiv 2021, posted 11 February. Full-text: https://doi.org/10.1101/2021.02.01.21250959
- DCGC 20210216. Genomic overview of SARS-CoV-2 in Denmark. Danish Covid-19 Genome Consortium 2021, published 16 February, accessed 20 February. Link: https://covid19genomics.dk/statistics
- De Souza WM, Amorm MR, Sesti-Costa R, et al. Levels of SARS-CoV-2 Lineage P.1 Neutralization by Antibodies Elicited after Natural Infection and Vaccination. Lancet Preprints 2021, posted 1 March. Full-text: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3793486
- Di Domenico L, Sabbatini CE, Pullano G, et al. Impact of January 2021 social distancing measures on SARS-CoV-2 B.1.1.7 circulation in France. INSERM 2021, published 14 February. Full-text: https://www.epicx-lab.com/uploads/9/6/9/4/9694133/inserm_covid-19-voc_socialdistancing-20210214.pdf
- Dong J, Zost SJ, Greaney AJ, et al. Genetic and structural basis for recognition of SARS-CoV-2 spike protein by a two-antibody cocktail. bioRxiv 2021, posted 28 January. Full-text: https://doi.org/10.1101/2021.01.27.428529
- du Plessis L, McCrone JT, Zarebski AE, et al. Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science 2021, published 8 January. Full-text: https://doi.org/10.1126/science.abf2946
- Du Z, Wang L, Yang B, et al. International risk of the new variant COVID-19 importations originating in the United Kingdom. medRxiv 2021, posted 12 January. Full-text: https://doi.org/10.1101/2021.01.09.21249384
- ECDC 20201220a. Threat Assessment Brief: Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom. European Centre for Disease Prevention and Control 2020, published 20 December. Full-text: https://www.ecdc.europa.eu/en/publications-data/threat-assessment-brief-rapid-increase-sars-cov-2-variant-united-kingdom
- ECDC 20201220b. Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom. European Centre for Disease Prevention and Control 2020, published 20 December. Full-text: https://www.ecdc.europa.eu/sites/default/files/documents/SARS-CoV-2-variant-multiple-spike-protein-mutations-United-Kingdom.pdf
- ECDC 20210121. Risk related to the spread of new SARS-CoV-2 variants of concern in the EU/EEA – first update. European Centre for Disease Prevention and Control 2021, published 21 January. Full-text: https://www.ecdc.europa.eu/en/publications-data/covid-19-risk-assessment-spread-new-variants-concern-eueea-first-update
- Emary KRW, Golubchik T, Aley PK, et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) Vaccine Against SARS-CoV-2 VOC 202012/01 (B.1.1.7). Lancet Preprints 2021, posted 4 February. Full-text: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3779160
- Faria NR, Claro IM, Candido D, et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological.org 2021, posted 12 January. Full-text: https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586
- Faria NR, Mellan TA, Whittaker C, et al. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. MedRxiv 2021, posted 3 March. Full-text: https://doi.org/10.1101/2021.02.26.21252554
- Fiocruz 20210129. Nota técnica da Fiocruz aborda Sars-CoV-2 e nova variante no AM. Fundação Oswaldo Cruz 2021, published 29 January. Full-text: https://portal.fiocruz.br/noticia/nota-tecnica-da-fiocruz-aborda-sars-cov-2-e-nova-variante-no-am
- Fiocruz. Frequencia das principais linhagens do sars-cov-2 por mes de amostragem. Fundação Oswaldo Cruz 2021. Link: http://www.genomahcov.fiocruz.br/frequencia-das-principais-linhagens-do-sars-cov-2-por-mes-de-amostragem/
- Fontanet A, Autran B, Lina B, et al. SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet 2021, published 11 February. Full-text: https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(21)00370-6/fulltext
- Fujino T, Nomoto H, Kutsuna S, et al. Novel SARS-CoV-2 Variant Identified in Travelers from Brazil to Japan. Emerg Infect Dis. 2021 Feb 10;27(4). PubMed: https://pubmed.gov/33567247. Full-text: https://doi.org/10.3201/eid2704.210138
- Galloway SE, Paul P, MacCannell DR, et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage — United States, December 29, 2020–January 12, 2021. MMWR Morb Mortal Wkly Rep. ePub: 15 January 2021. Full-text: http://dx.doi.org/10.15585/mmwr.mm7003e2
- Garcia-Beltran WF, Lam EC, St. Denis K, et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021, 12 March. Full-text: https://www.cell.com/cell/fulltext/S0092-8674(21)00298-1
- Gaymard A, Bosetti P, Feri A, et al. Early assessment of diffusion and possible expansion of SARS-CoV-2 Lineage 20I/501Y.V1 (B.1.1.7, variant of concern 202012/01) in France, January to March 2021. Euro Surveill. 2021 Mar;26(9). PubMed: https://pubmed.gov/33663644. Full-text: https://doi.org/10.2807/1560-7917.ES.2021.26.9.2100133
- Gottlieb RL, Nirula A, Chen P, et al. Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA. 2021 Feb 16;325(7):632-644. PubMed: https://pubmed.gov/33475701. Full-text: https://doi.org/10.1001/jama.2021.0202
- Gozlan M. Covid-19 : le défi des nouveaux variants. Réalités Biomédicales 2021, published 18 January. Full-text : https://www.lemonde.fr/blog/realitesbiomedicales/2021/01/18/covid-19-le-defi-des-nouveaux-variants/
- Gravagnuolo AM, Faqih L, Cronshaw C, et al. Epidemiological Investigation of New SARS-CoV-2 Variant of Concern 202012/01 in England. medRxiv 2021, posted 15 January. Full-text: https://doi.org/10.1101/2021.01.14.21249386
- Greaney AJ, Loes AN, Crawford KHD, et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021 Mar 10;29(3):463-476.e6. PubMed: https://pubmed.gov/33592168. Full-text: https://doi.org/10.1016/j.chom.2021.02.003
- Greaney AJ, Starr TN, Gilchuk P, et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe. 2021 Jan 13;29(1):44-57.e9. PubMed: https://pubmed.gov/33259788. Full-text: https://doi.org/10.1016/j.chom.2020.11.007
- Grint DJ, Wing K, Williamson E, et al. Case fatality risk of the SARS-CoV-2 variant of concern B.1.1.7 in England, 16 November to 5 February. Volume 26, Issue 11, 18 March 2021. https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2021.26.11.2100256
- Gu H, Chen Q, Yang G, et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020 Sep 25;369(6511):1603-1607. PubMed: https://pubmed.gov/32732280. Full-text: https://doi.org/10.1126/science.abc4730
- Gupta V, Bhoyar RC, Jain A, et al. Asymptomatic reinfection in two healthcare workers from India with genetically distinct SARS-CoV-2. Clin Infect Dis. 2020 Sep 23:ciaa1451. PubMed: https://pubmed.gov/32964927. Full-text: https://doi.org/10.1093/cid/ciaa1451
- Hadfield J, Megill C, Bell SM, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018 Dec 1;34(23):4121-4123. PubMed: https://pubmed.gov/29790939. Full-text: https://doi.org/10.1093/bioinformatics/bty407
- Hall VJ, Foulkes S, Saei A, et al. Effectiveness of BNT162b2 mRNA Vaccine Against Infection and COVID-19 Vaccine Coverage in Healthcare Workers in England, Multicentre Prospective Cohort Study (the SIREN Study) Victoria Jane Hall. Lancet Preprints 2021, published 22 February. Full-text: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3790399
- Hansen J, Baum A, Pascal KE, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020 Aug 21;369(6506):1010-1014. PubMed: https://pubmed.gov/32540901. Full-text: https://doi.org/10.1126/science.abd0827
- Haynes WA, Kamath K, Lucas C, Shon J, Iwasaki A. Impact of B.1.1.7 variant mutations on antibody recognition of linear SARS-CoV-2 epitopes. medRxiv 2021, posted 8 January. Full-text: https://doi.org/10.1101/2021.01.06.20248960
- Helix 202102. The Helix® COVID-19 Surveillance Dashboard. Helix 2021; website: https://www.helix.com/pages/helix-covid-19-surveillance-dashboard
- Hensley MK, Bain WG, Jacobs J, et al. Intractable Coronavirus Disease 2019 (COVID-19) and Prolonged Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Replication in a Chimeric Antigen Receptor-Modified T-Cell Therapy Recipient: A Case Study. Clin Infect Dis 2021. Full-text: https://doi.org/10.1093/cid/ciab072
- Hodcroft EB, Domman DB, Snyder DJ, et al. Emergence in late 2020 of multiple lineages of SARS-CoV-2 Spike protein variants affecting amino acid position 677. medRxiv 2021, posted 14 January. Full-text: https://doi.org/10.1101/2021.02.12.21251658
- Hoffmann M, Arora P, Groß R, et al. SARS-CoV-2 variants B.1.351 and B.1.1.248: Escape from therapeutic antibodies and antibodies induced by infection and vaccination. medRxiv 2021, posted 11 February. Full-text: https://doi.org/10.1101/2021.02.11.430787
- Hoffmann M, Kleine-Weber H, Pöhlmann S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol Cell. 2020 May 21;78(4):779-784.e5. PubMed: https://pubmed.gov/32362314. Full-text: https://doi.org/10.1016/j.molcel.2020.04.022
- Horby P, Huntley C, Davies N, Edmunds J, Ferguson N, Medley G, Hayward A, Cevik M, Semple C. NERVTAG note on B.1.1.7 severity. SAGE meeting date 21 January. Full-text: https://bit.ly/3o93gYP
- INSA 20210112 – Instituto Nacional de Saúde Doutor Ricardo Jorge. Diversidade genética do novo coronavírus SARS-CoV-2 (COVID-19) em Portugal – Relatório de situação [updated 12 January 2021; accessed 26 January 2021]. Full-text: https://insaflu.insa.pt/covid19/relatorios/INSA_SARS_CoV_2_DIVERSIDADE_GENETICA_relatorio_situacao_2021-01-12.pdf
- Iwasaki A. What reinfections mean for COVID-19. Lancet Infect Dis. 2021 Jan;21(1):3-5. PubMed: https://pubmed.gov/33058796. Full-text: https://doi.org/10.1016/S1473-3099(20)30783-0
- Jangra S, Ye C, Rathnasinghe R, et al. The E484K mutation in the SARS-CoV-2 spike protein reduces but does not abolish neutralizing activity of human convalescent and post-vaccination sera. medRxiv. 2021 Jan 29:2021.01.26.21250543. PubMed: https://pubmed.gov/33532796. Full-text: https://doi.org/10.1101/2021.01.26.21250543
- JNJ 20210129. Johnson & Johnson Announces Single-Shot Janssen COVID-19 Vaccine Candidate Met Primary Endpoints in Interim Analysis of its Phase 3 ENSEMBLE Trial. Johnson & Johnson 2021, published 29 January. Full-text: https://www.jnj.com/johnson-johnson-announces-single-shot-janssen-covid-19-vaccine-candidate-met-primary-endpoints-in-interim-analysis-of-its-phase-3-ensemble-trial
- Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020 Aug;584(7819):115-119. PubMed: https://pubmed.gov/32454513. Full-text: https://doi.org/10.1038/s41586-020-2380-z
- Kemp SA, Collier DA, Datir R, et al. Neutralising antibodies in Spike mediated SARS-CoV-2 adaptation. medRxiv. 2020 Dec 29:2020.12.05.20241927. PubMed: https://pubmed.gov/33398302. Full-text: https://doi.org/10.1101/2020.12.05.20241927
- Kirby T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir Med 2021, published 5 January. Full-text: https://doi.org/10.1016/S2213-2600(21)00005-9
- Kissler S, Fauver JR, Mack C, et al. Densely sampled viral trajectories suggest longer duration of acute infection with B.1.1.7 variant relative to non-B.1.1.7 SARS-CoV-2. medRxiv 2021, posted 19 February. Full-text: https://www.medrxiv.org/content/10.1101/2021.02.16.21251535v1
- Kistler KE, Bedford T. Evidence for adaptive evolution in the receptor-binding domain of seasonal coronaviruses OC43 and 229e. Elife. 2021 Jan 19;10:e64509. PubMed: https://pubmed.gov/33463525. Full-text: https://doi.org/10.7554/eLife.64509
- Koff WC, Berkley SF. A universal coronavirus vaccine. Science 2021, published 19 February. Full-text: https://doi.org/10.1126/science.abh0447
- Korber B, Fischer WM, Gnanakaran S, et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020 Aug 20;182(4):812-827.e19. PubMed: https://pubmed.gov/32697968. Full-text: https://doi.org/10.1016/j.cell.2020.06.043
- Kreer C, Zehner M, Weber T, et al. Longitudinal Isolation of Potent Near-Germline SARS-CoV-2-Neutralizing Antibodies from COVID-19 Patients. Cell. 2020 Sep 17;182(6):1663-1673. PubMed: https://pubmed.gov/32946786. Full-text: https://doi.org/10.1016/j.cell.2020.08.046
- Kupferschmidt 20201220. Mutant coronavirus in the United Kingdom sets off alarms, but its importance remains unclear. Nature 2020, published 20 December. Full-text: https://www.sciencemag.org/news/2020/12/mutant-coronavirus-united-kingdom-sets-alarms-its-importance-remains-unclear
- Kupferschmidt 20201223. U.K. variant puts spotlight on immunocompromised patients’ role in the COVID-19 pandemic. Nature 2020, published 23 December. Full-text: https://www.sciencemag.org/news/2020/12/uk-variant-puts-spotlight-immunocompromised-patients-role-covid-19-pandemic
- Kupferschmidt 20210151. New coronavirus variants could cause more reinfections, require updated vaccines. Nature 2021, published 15 January. Full-text: https://www.sciencemag.org/news/2021/01/new-coronavirus-variants-could-cause-more-reinfections-require-updated-vaccines
- Kupferschmidt 20210203. Danish scientists see tough times ahead as they watch more contagious COVID-19 virus surge. Science 2021, published 3 February. Full-text: https://www.sciencemag.org/news/2021/02/danish-scientists-see-tough-times-ahead-they-watch-more-contagious-covid-19-virus-surge
- Larson D, Brodniak SL, Voegtly LJ, et al. A Case of Early Re-infection with SARS-CoV-2. Clin Infect Dis. 2020 Sep 19:ciaa1436. PubMed: https://pubmed.gov/32949240. Full-text: https://doi.org/10.1093/cid/ciaa1436ed.gov/32950102
- Lauring AS, Hodcroft EB. Genetic Variants of SARS-CoV-2—What Do They Mean? JAMA 2021, published 6 January. Full-text: https://doi.org/10.1001/jama.2020.27124
- Le Monde 20210126. Covid-19 : le variant anglais représente près d’un cas positif sur dix en Ile-de-France. Le Monde, published 26 January. Full-text : https://www.lemonde.fr/planete/article/2021/01/26/covid-19-une-pme-sur-deux-craint-de-ne-pas-pouvoir-supporter-un-troisieme-confinement_6067608_3244.html
- Leach A, Kirk A, Duncan P. ‘Encouraging’ signs for Covid vaccine as over-80s deaths fall in England. The Guardian 2021, published 16 February. Full-text: https://www.theguardian.com/society/2021/feb/16/encouraging-signs-covid-vaccine-over-80s-deaths-fall-england
- Ledford H 20210114. COVID reinfections are unusual — but could still help the virus to spread. Nature 2021, published 14 January. Full-text: https://www.nature.com/articles/d41586-021-00071-6
- Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021 Jan;26(1). PubMed: https://pubmed.gov/33413740. Full-text: https://doi.org/10.2807/1560-7917.ES.2020.26.1.2002106
- Levine-Tiefenbrun M, Yelin I, Katz R, et al. Decreased SARS-CoV-2 viral load following vaccination. MedRxiv 2021, posted 8 February. Full-text: https://www.medrxiv.org/content/10.1101/2021.02.06.21251283v1
- Liu H, Wu NC, Yuan M, et al. Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity. Immunity. 2020 Dec 15;53(6):1272-1280.e5. PubMed: https://pubmed.gov/33242394. Full-text: https://doi.org/10.1016/j.immuni.2020.10.023
- Liu L, Wang P, Nair MS, et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020 Aug;584(7821):450-456. PubMed: https://pubmed.gov/32698192. Full-text: https://doi.org/10.1038/s41586-020-2571-7
- Liu Y, Liu J, Xia H, et al. Neutralizing Activity of BNT162b2-Elicited Serum – Preliminary Report. N Engl J Med. 2021 Feb 17. PubMed: https://pubmed.gov/33596352. Full-text: https://doi.org/10.1056/NEJMc2102017
- Liu Z, VanBlargan LA, Bloyet LM, et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021 Mar 10;29(3):477-488.e4. PubMed: https://pubmed.gov/33535027. Full-text: https://doi.org/10.1016/j.chom.2021.01.014
- Liu Z, VanBlargan LA, Bloyet LM, et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 2021, published 27 January. Full-text: https://doi.org/10.1016/j.chom.2021.01.014
- Madhi SA, Baillie V, Cutland CL, et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N Engl J Med. 2021 Mar 16. PubMed: https://pubmed.gov/33725432. Full-text: https://doi.org/10.1056/NEJMoa2102214
- Maggi F, Novazzi F, Genoni A, et al. Imported SARS-COV-2 Variant P.1 Detected in Traveler Returning from Brazil to Italy. Emerg Infect Dis. 2021 Feb 10;27(4). PubMed: https://pubmed.gov/33567246. Full-text: https://doi.org/10.3201/eid2704.210183
- Mahase E. Covid-19: What new variants are emerging and how are they being investigated? BMJ. 2021 Jan 18;372:n158. PubMed: https://pubmed.gov/33462092. Full-text: https://doi.org/10.1136/bmj.n158
- Mahdi SA, Baillie C, Cutland CL, et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. NEJM, March 16, 2021. https://www.nejm.org/doi/full/10.1056/NEJMoa2102214
- Mallapaty S. Can COVID vaccines stop transmission? Scientists race to find answers. Nature 2021, published 19 February. Full-text: https://www.nature.com/articles/d41586-021-00450-z
- Mandavilli A. Immunity to the Coronavirus May Last Years, New Data Hint. The New York Times 2020, published 17 November. Full-text: https://www.nytimes.com/2020/11/17/health/coronavirus-immunity.html
- McCallum M, Marco A, Lempp F, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. bioRxiv. 2021 Jan 14:2021.01.14.426475. PubMed: https://pubmed.gov/33469588. Full-text: https://doi.org/10.1101/2021.01.14.426475
- McCarthy KR, Rennick LJ, Namnulli S, et al. Natural deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. bioRxiv 2020, posted 19 November. Full-text: https://www.biorxiv.org/content/10.1101/2020.11.19.389916v1
- Moderna 20210125. Moderna COVID-19 Vaccine Retains Neutralizing Activity Against Emerging Variants First Identified in the U.K. and the Republic of South Africa. Moderna 2021, published 25 January, accessed 29 January. Full-text: https://investors.modernatx.com/news-releases/news-release-details/moderna-covid-19-vaccine-retains-neutralizing-activity-against
- Moore S, Hill EM, Tildesley MJ, Dyson L, Keeling MJ. Vaccination and non-pharmaceutical interventions for COVID-19: a mathematical modelling study. Lancet Infect Dis 2021, published 18 March. Full-text: https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(21)00143-2/fulltext
- Moyo-Gwete T, Madzivhandila M, Makhado Z, et al. SARS-CoV-2 501Y.V2 (B.1.351) elicits cross-reactive neutralizing antibodies. bioRxiv 2021, posted 6 March. Full-text: https://doi.org/10.1101/2021.03.06.434193
- Muik A, Wallisch AK, Sänger B, et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine–elicited human sera. Science 2021, published 29 January. Full-text: https://doi.org/10.1126/science.abg6105
- MV 20210131. 98e OMT advies deel 1. Ministerie van Volksgezondheid et al. 2021, published 31 January. Full-text: https://www.rijksoverheid.nl/documenten/rapporten/2021/01/31/98e-omt-advies-deel-1
- Naveca F, da Costa C, Nascimento V, et al. SARS-CoV-2 reinfection by the new Variant of Concern (VOC) P.1 in Amazonas, Brazil. Virological 2021, posted 18 January. Full-text: https://virological.org/t/sars-cov-2-reinfection-by-the-new-variant-of-concern-voc-p-1-in-amazonas-brazil/596
- Naveca F, Nascimento V, Souza V, et al. COVID-19 epidemic in the Brazilian state of Amazonas was driven by long-term persistence of endemic SARS-CoV-2 lineages and the recent emergence of the new Variant of Concern P.1. Research Square 2021, posted 25 February. Full-text: https://doi.org/10.21203/rs.3.rs-275494/v1
- Neidleman J, Luo X, Frouard J, et al. SARS-CoV-2-Specific T Cells Exhibit Phenotypic Features of Helper Function, Lack of Terminal Differentiation, and High Proliferation Potential. Cell Rep Med. 2020 Sep 22;1(6):100081. PubMed: https://pubmed.gov/32839763. Full-text: https://doi.org/10.1016/j.xcrm.2020.100081
- NERVTAG 20210211. Update note on B.1.1.7 severity. New and Emerging Respiratory Virus Threats Advisory Group (NERVTAG) 2021, published 12 February. Full-text: https://www.gov.uk/government/publications/nervtag-update-note-on-b117-severity-11-february-2021
- Novavax 20210128. Novavax COVID-19 Vaccine Demonstrates 89.3% Efficacy in UK Phase 3 TrialNovavax COVID-19 Vaccine Demonstrates 89.3% Efficacy in UK Phase 3 Trial. Novavax 2021, published 28 January, accessed 30 January. Full-text: https://ir.novavax.com/news-releases/news-release-details/novavax-covid-19-vaccine-demonstrates-893-efficacy-uk-phase-3
- Novavax 20210311. Novavax Confirms High Levels of Efficacy Against Original and Variant COVID-19 Strains in United Kingdom and South Africa Trials. Novavax 2021, 11 March press release. Full-text: https://ir.novavax.com/news-releases/news-release-details/novavax-confirms-high-levels-efficacy-against-original-and-0
- Pearson CAB, Russel TW, Davies N, Kucharski AJ. Estimates of severity and transmissibility of novel South Africa SARS-CoV-2 variant 501Y.V2. CMMID Repository 2021, published 11 January. Full-text: https://cmmid.github.io/topics/covid19/sa-novel-variant.html
- Petter E, Mor O, Zuckermann N, et al. Initial real world evidence for lower viral load of individuals who have been vaccinated by BNT162b2. GitHub 2021, posted 7 February. Full-text: http://bit.ly/3aPIetS
- Piccoli L, Park YJ, Tortorici MA, et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell. 2020 Nov 12;183(4):1024-1042.e21. PubMed: https://pubmed.gov/32991844. Full-text: https://doi.org/10.1016/j.cell.2020.09.037
- Pinto D, Park YJ, Beltramello M, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020 Jul;583(7815):290-295. PubMed: https://pubmed.gov/32422645. Full-text: https://doi.org/10.1038/s41586-020-2349-y
- Plante JA, Liu Y, Liu J, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 Oct 26. PubMed: https://pubmed.gov/33106671. Full-text: https://doi.org/10.1038/s41586-020-2895-3
- Plante JA, Mitchell BM, Plante KS, et al. The Variant Gambit: COVID’s Next Move. Cell Host Microbe March 01, 2021. https://www.cell.com/cell-host-microbe/fulltext/S1931-3128(21)00099-8
- Prévost J, Finzi A. The great escape? SARS-CoV-2 variants evading neutralizing responses. Cell Host Microbe. 2021 Mar 10;29(3):322-324. PubMed: https://pubmed.gov/33705702. Full-text: https://doi.org/10.1016/j.chom.2021.02.010
- Priesemann V, Balling R, Brinkmann MM, et al. An action plan for pan-European defence against new SARS-CoV-2 variants. Lancet. 2021 Jan 21:S0140-6736(21)00150-1. PubMed: https://pubmed.gov/33485462. Full-text: https://doi.org/10.1016/S0140-6736(21)00150-1
- Public Health England 20201228. Investigation of novel SARS-COV-2 variant: Variant of Concern 202012/01 – Technical briefing 2. UK Government 2020, updated 28 December; accessed 22 January 2021. Full-text: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/949639/Technical_Briefing_VOC202012-2_Briefing_2_FINAL.pdf
- Public Health England 20210105. Investigation of novel SARS-COV-2 variant: Variant of Concern 202012/01 – Technical briefing 3. UK Government 2021, updated 5 January; accessed 22 January 2021. Full-text: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/950823/Variant_of_Concern_VOC_202012_01_Technical_Briefing_3_-_England.pdf
- Public Health England 20210115. Investigation of novel SARS-COV-2 variant: Variant of Concern 202012/01 – Technical briefing 4. UK Government 2021, updated 15 January; accessed 22 January 2021. Full-text: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/952490/Variant_of_Concern_VOC_202012_01_Technical_Briefing_4_England.pdf
- Public Health England 20210126. Investigation of novel SARS-COV-2 variant: Variant of Concern 202012/01 – Technical briefing 5. UK Government 2021, updated 26 January; accessed 2 February 2021. Full-text: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/957504/Variant_of_Concern_VOC_202012_01_Technical_Briefing_5_England.pdf
- Public Health England 20210222. PHE monitoring of the early impact and effectiveness of COVID-19 vaccination in England. UK Government 2021, 22 February; accessed 5 March 2021. Full-text: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/963532/COVID-19_vaccine_effectiveness_surveillance_report_February_2021_FINAL.pdf
- Rambaut A, Holmes EC, O’Toole Á, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020b Nov;5(11):1403-1407. PubMed: https://pubmed.gov/32669681. Full-text: https://doi.org/10.1038/s41564-020-0770-5
- Rambaut A, Loman N, Pybus O, et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological 2020, Full-text: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563/1
- Resende PC, Bezerra JF, Teixeira de Vasconcelos RH, et al. Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil, 2020. Virological.org 2020, posted December. Full-text: https://virological.org/t/spike-e484k-mutation-in-the-first-sars-cov-2-reinfection-case-confirmed-in-brazil-2020/584/1
- RKI 20210205. Bericht zu Virusvarianten von SARS-CoV-2 in Deutschland, insbesondere zur Variant of Concern (VOC) B.1.1.7 (5.2.2021). Robert Koch Institut 2021, published 5 February 2021, accessed 5 February 2021. Full-text: https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/DESH/Bericht_VOC_05022021
- Robbiani DF, Gaebler C, Muecksch F, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020 Aug;584(7821):437-442. PubMed: https://pubmed.gov/32555388. Full-text: https://doi.org/10.1038/s41586-020-2456-9
- Rogers TF, Zhao F, Huang D, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020 Aug 21;369(6506):956-963. PubMed: https://pubmed.gov/32540903. Full-text: https://doi.org/10.1126/science.abc7520
- Rossman H, Shilo S, Meir T, Gorfine M,Shalit U, Segal E. Patterns of covid-19 pandemic dynamics following deployment of a broad national immunization program. GitHub 2021, posted 3 February. Full-text: http://bit.ly/36KhjOU
- Sabino EC, Buss LF, Carvalho MPS, et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet 2021, published 27 January. Full-text: https://doi.org/10.1016/S0140-6736(21)00183-5
- Sampedro J. El evento de Ischgl. El País 2020, published 3 October. Full-text: https://elpais.com/ciencia/2020-10-02/el-evento-de-ischgl.html
- Schultze JL, Aschenbrenner AC. COVID-19 and the human innate immune system. Cell 2021, published 16 February. Full-text: https://www.cell.com/cell/fulltext/S0092-8674(21)00218-X
- Segal E, Topol EJ, Verghese A. The Impact of the Vaccine Has Been Tremendous. Medscape 2021, 17 February. Accessed 17 February. Audio and transcript: https://www.medscape.com/viewarticle/945680
- Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021 Feb 18;184(4):861-880. PubMed: https://pubmed.gov/33497610. Full-text: https://doi.org/10.1016/j.cell.2021.01.007
- Shi R, Shan C, Duan X, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020 Aug;584(7819):120-124. PubMed: https://pubmed.gov/32454512. Full-text: https://doi.org/10.1038/s41586-020-2381-y
- Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 2017 Mar 30;22(13):30494. PubMed: https://pubmed.gov/28382917. Full-text: https://doi.org/10.2807/1560-7917.ES.2017.22.13.30494
- Skelly DT, Harding AC, Gilbert Jaramillo J, et al. Vaccine-induced immunity provides more robust heterotypic immunity than natural infection to emerging SARS-CoV-2 variants of concern. Research Square 2021, posted 9 February, accessed 12 February. Full-text: https://www.researchsquare.com/article/rs-226857/v1?s=09
- South African Government 20201218. Minister Zweli Mkhize confirms 8 725 more cases of Coronavirus COVID-19. South African Government 2020, published 18 December 2020; accessed 28 January 2021. Full-text: https://www.gov.za/speeches/minister-zweli-mkhize-confirms-8-725-more-cases-coronavirus-covid-19-18-dec-2020-0000
- SSI 20210203. Status for udvikling af B.1.1.7 og andre mere smitsomme varianter i Danmark. Statens Serum Institut 2021, published 3 February, accessed 4 February. Full-text: https://files.ssi.dk/covid19/virusvarianter/status/status-virusvarianter-03022021-q75m
- SSI 20210204. Kontakttal for virusvariant B.1.1.7. Statens Serum Institut 2021, published 4 February, accessed 6 February. Full-text: https://www.ssi.dk/aktuelt/nyheder/2021/kontakttal-for-virusvariant-B.1.1.7
- SSI 20210209. Kontakttal for B.1.1.7 og andre hyppige virusvarianter. Statens Serum Institut 2021, published 9 February, accessed 11 February. Full-text: https://www.ssi.dk/-/media/cdn/files/kontakttal-for-B.1.1.7-og-andre-hyppige-virusvarianter0902221.pdf
- Stamatatos L, Czartoski J, Wan YH, et al. Antibodies elicited by SARS-CoV-2 infection and boosted by vaccination neutralize an emerging variant and SARS-CoV-1. MedRxiv 2021, posted 8 February. Full-text: https://www.medrxiv.org/content/10.1101/2021.02.05.21251182v1
- Starr TN, Greaney AJ, Addetia A, et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 2021, published 25 January. Full-text: https://doi.org/10.1126/science.abf9302
- Starr TN, Greaney AJ, Dingens AS, Bloom JD. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. bioRxiv 2021, posted 21 February. Full-text: https://doi.org/10.1101/2021.02.17.431683
- Starr TN, Greaney AJ, Hilton SK, et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020 Sep 3;182(5):1295-1310.e20. PubMed: https://pubmed.gov/32841599. Full-text: https://doi.org/10.1016/j.cell.2020.08.012
- Stromboni C. Sous la pression du variant anglais, Dunkerque voit flamber l’épidémie de Covid-19. Le Monde 2021, published 20 February. Full-text : https://www.lemonde.fr/planete/article/2021/02/20/sous-la-pression-du-variant-anglais-dunkerque-voit-flamber-l-epidemie-de-covid-19_6070631_3244.html
- Supasa P, Daming Z, Dejnirattisai W, et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell 2021, published 18 February. Full-text: https://www.cell.com/cell/fulltext/S0092-8674(21)00222-1
- Swiss National COVID-19 Taskforce. SARS-CoV-2 Variants of Concern in Switzerland. Updated 9 February. Links: https://sciencetaskforce.ch/en/nextstrain-phylogenetic-analysis/ + https://ibz-shiny.ethz.ch/covidDashboard/variant-plot/index.html
- Tarke A, Sidney J, Methot N, et al. Negligible impact of SARS-CoV-2 variants on CD4 + and CD8 + T cell reactivity in COVID-19 exposed donors and vaccinees. bioRxiv. 2021 Mar 1:2021.02.27.433180. PubMed: https://pubmed.gov/33688655. Full-text: https://doi.org/10.1101/2021.02.27.433180
- Tegally H, Wilkinson E, Giovanetti M, et al. Emergence of a SARS-CoV-2 variant of concern with mutations in spike glycoprotein. Nature. 2021 Mar 9. PubMed: https://pubmed.gov/33690265. Full-text: https://doi.org/10.1038/s41586-021-03402-9
- Thomson EC, Rosen LE, Shepherd JG, et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021 Mar 4;184(5):1171-1187.e20. PubMed: https://pubmed.gov/33621484. Full-text: https://doi.org/10.1016/j.cell.2021.01.037
- Tillett RL, Sevinsky JR, Hartley PD, et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2021 Jan;21(1):52-58. PubMed: https://pubmed.gov/33058797. Full-text: https://doi.org/10.1016/S1473-3099(20)30764-7
- To KK, Hung IF, Ip JD, et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2020 Aug 25:ciaa1275. PubMed: https://pubmed.gov/32840608. Full-text: https://doi.org/10.1093/cid/ciaa1275
- Tufekci Z. The Mutated Virus Is a Ticking Time Bomb. The Atlantic 2020, published 31 December. Full-text: https://www.theatlantic.com/science/archive/2020/12/virus-mutation-catastrophe/617531
- Tzou PL, Tao K, Nouhin J, et al. Coronavirus Antiviral Research Database (CoV-RDB): An Online Database Designed to Facilitate Comparisons between Candidate Anti-Coronavirus Compounds. Viruses. 2020 Sep 9;12(9):1006. PubMed: https://pubmed.gov/32916958. Full-text: https://doi.org/10.3390/v12091006
- Van Elslande J, Vermeersch P, Vandervoort K, et al. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin Infect Dis. 2020 Sep 5:ciaa1330. PubMed: https://pubmed.gov/32887979. Full-text: https://doi.org/10.1093/cid/ciaa1330
- Vasileiou E, Simpson CR, Robertson C, et al. Effectiveness of First Dose of COVID-19 Vaccines Against Hospital Admissions in Scotland: National Prospective Cohort Study of 5.4 Million People. Lancet Preprints 2021, posted 19 February. Full-text: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3789264
- Vasques Nonaka CK, Franco M, Gräf T, et al. Genomic Evidence of a Sars-Cov-2 Reinfection Case With E484K Spike Mutation in Brazil. Preprints 2021, posted 6 January. Full-text: https://doi.org/10.20944/preprints202101.0132.v1
- Voloch CM, da Silva R, de Almeida LGP, et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. medRxiv 2020, posted 26 December. Full-text: https://doi.org/10.1101/2020.12.23.20248598
- Volz E, Hill V, McCrone JT, et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell. 2021a Jan 7;184(1):64-75.e11. PubMed: https://pubmed.gov/33275900. Full-text: https://doi.org/10.1016/j.cell.2020.11.020
- Volz E, Mishra W, Chand M, et al. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. medRxiv 2021, posted 4 January. Full-text: https://doi.org/10.1101/2020.12.30.20249034
- Wang P, Liu L, Nair MS, et al. SARS-CoV-2 neutralizing antibody responses are more robust in patients with severe disease. Emerg Microbes Infect. 2020 Dec;9(1):2091-2093. PubMed: https://pubmed.gov/32930052. Full-text: https://doi.org/10.1080/22221751.2020.1823890
- Wang P, Nair MS, Liu L, et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature March 8, 2021. https://www.nature.com/articles/s41586-021-03398-2
- Wang P, Wang M, Yu J, et al. Increased Resistance of SARS-CoV-2 Variant P.1 to Antibody Neutralization. bioRxiv 2021, posted 2 March. Full-text: https://doi.org/10.1101/2021.03.01.433466
- Wang Z, Schmidt F, Weisblum Y, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021, published 10 February. Full-text: https://doi.org/10.1101/2021.01.15.426911
- Washington NL, Gangavarapu K, Zeller M, et al. Genomic epidemiology identifies emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. medRxiv 2021, posted 7 February. Full-text: https://www.medrxiv.org/content/10.1101/2021.02.06.21251159v1
- Weinreich DM, Sivapalasingam S, Norton T, et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. NEJM December 17, 2020, Full-text: https://doi.org/10.1056/NEJMoa2035002
- Weisblum Y, Schmidt F, Zhang F, et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020 Oct 28;9:e61312. PubMed: https://pubmed.gov/33112236. Full-text: https://doi.org/10.7554/eLife.61312
- WHO 20201231. SARS-CoV-2 variants. WHO 2020, published 31 December. Full-text: https://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en/
- Wibmer CK, Ayres F, Hermanus T, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021 Mar 2. PubMed: https://pubmed.gov/33654292. Full-text: https://doi.org/10.1038/s41591-021-01285-x
- Wise J. Covid-19: The E484K mutation and the risks it poses. BMJ. 2021 Feb 5;372:n359. PubMed: https://pubmed.gov/33547053. Full-text: https://doi.org/10.1136/bmj.n359
- Wong AHM, Tomlinson ACA, Zhou D, et al. Receptor-binding loops in alphacoronavirus adaptation and evolution. Nat Commun. 2017 Nov 23;8(1):1735. PubMed: https://pubmed.gov/29170370. Full-text: https://doi.org/10.1038/s41467-017-01706-x
- Wu K, Werner AP, Koch M, et al. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine – Preliminary Report. N Engl J Med. 2021 Feb 17. PubMed: https://pubmed.gov/33596346. Full-text: https://doi.org/10.1056/NEJMc2102179
- Xie X, Liu Y, Liu J, et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med 2021, published 8 February. Full-text: https://www.nature.com/articles/s41591-021-01270-4
- Xie X, Zou J, Fonte-Garfias CR, et al. Neutralization of N501Y mutant SARS-CoV-2 by BNT162b2 vaccine-elicited sera. bioRxiv 2021, posted 7 January. Full-text: https://doi.org/10.1101/2021.01.07.425740
- Yurkovetskiy L, Wang X, Pascal KE, et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020 Oct 29;183(3):739-751.e8. PubMed: https://pubmed.gov/32991842. Full-text: https://doi.org/10.1016/j.cell.2020.09.032
- Zahradnik J, Marciano S, Shemesh M, et al. SARS-CoV-2 RBD in vitro evolution follows contagious mutation spread, yet generates an able infection inhibitor. bioRxiv 2021, posted 8 January. Full-text: https://www.biorxiv.org/content/10.1101/2021.01.06.425392v2
- Zhang W, Davis BD, Chen SS, Sincuir Martinez JM, Plummer JT, Vail E. Emergence of a Novel SARS-CoV-2 Variant in Southern California. JAMA. 2021 Feb 11. PubMed: https://pubmed.gov/33571356. Full-text: https://doi.org/10.1001/jama.2021.1612
- Zimmer C. Virus Variant in Brazil Infected Many Who Had Already Recovered From Covid-19. The New York Times 2021, published 3 March. Full-text: https://www.nytimes.com/2021/03/01/health/covid-19-coronavirus-brazil-variant.html
- Zost SJ, Gilchuk P, Case JB, et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020 Aug;584(7821):443-449. PubMed: https://pubmed.gov/32668443. Full-text: https://doi.org/10.1038/s41586-020-2548-6
