24 November
Bushman M, Kahn R, Taylor BP, et al. Population impact of SARS-CoV-2 variants with enhanced transmissibility and/or partial immune escape. Cell November 18, 2021. https://www.cell.com/cell/fulltext/S0092-8674(21)01374-X
Mathematical modeling study, showing that variants with enhanced transmissibility frequently increase epidemic severity, whereas those with partial immune escape either fail to spread widely, or primarily cause breakthrough infections. However, when these phenotypes are combined, a variant can continue spreading even as immunity builds up in the population, limiting the impact of vaccination and exacerbating the epidemic. The findings are consistent with the global sweeps by highly transmissible variants such as Alpha and Delta, as well as the failure of Beta to reach high frequency in most areas.
15 November
Zuckerman N, Nemet I, Kliker L, et al. The SARS-CoV-2 Lambda variant and its neutralisation efficiency following vaccination with Comirnaty, Israel, April to June 2021. Euro Surveill November 11, 2021; 26(45):pii=2100974. https://doi.org/10.2807/1560-7917.ES.2021.26.45.2100974
The Lambda variant of interest (VOI) was first detected in Lima, Peru in August 2020. By April 2021, the proportion reached nearly 100% of sequenced genomic isolates detected in Peru. Its spread in South America occurred despite the presence of additional lineages, including Alpha and Gamma. In this study, neutralization assays of the Lambda VOI with sera from 36 individuals vaccinated with two doses of the Comirnaty vaccine demonstrated a subtle yet statistically significant 1.6-fold decrease in neutralization capacity of the Lambda variant compared with the WT SARS-CoV-2 strain. Thus, the vaccine efficacy against the Lambda VOI was similarly compromised as compared with the Delta VOI.
12 November
McCallum M, Walls AC, Sprouse KR, et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science, November 2021. https://www.science.org/doi/10.1126/science.abl8506
In late 2020, B.1.617 variants including B.1.617.1 (Kappa) and B.1.617.2 (Delta) were first detected in India. Mutations in the Kappa and Delta spike glycoproteins abrogate recognition by several monoclonal antibodies via alteration of key antigenic sites, including remodeling of the Delta N-terminal domain.
11 November
de Gier B, Stijn A, Backer JA, et al. Vaccine effectiveness against SARS-CoV-2 transmission to household contacts during dominance of Delta variant (B.1.617.2), the Netherlands, August to September 2021. Euro Surveill. 2021;26(44):pii=2100977. https://doi.org/10.2807/1560-7917.ES.2021.26.44.2100977
Vaccination confers protection against onward transmission of SARS-CoV-2 from vaccinated index cases, albeit somewhat less for the Delta than for the Alpha variant.
7 November
Uriu K, Kimura I, Shirakawa K, et al. Neutralization of the SARS-CoV-2 Mu Variant by Convalescent and Vaccine Serum. NEJM November 2021. https://www.nejm.org/doi/full/10.1056/NEJMc2114706?query=featured_home
Mu (B.1.621) represents the most recently recognized variant of interest. The epicenter of mu transmission is Colombia, where the variant has outnumbered all other variants, including Gamma. This work shows that mu has a pronounced resistance to antibodies elicited by natural SARS-CoV-2 infection and by the BNT162b2 mRNA vaccine.
1 November
Horiuchi S, Oishi K, Carrau L, et al. Immune memory from SARS-CoV-2 infection in hamsters provides variant-independent protection but still allows virus transmission. Science Immunology October 26, 2021. https://www.science.org/doi/10.1126/sciimmunol.abm3131
Re-infection of recovered animals with a SARS-CoV-2 variant of concern showed that SARS-CoV-2-specific T and B cells could effectively control the infection associated with the rapid induction of neutralizing antibodies but failed to block transmission to both naïve and seroconverted animals. (It is expected that Joshua may conclude that “hamsters are not humans”).
Kant R, Nguyen PT, Blomqvist S, et al. Incidence trends for SARS-CoV-2 Alpha and Beta variants, Finland, spring 2021. Emerg Infect Dis. 2021 October 27. https://wwwnc.cdc.gov/eid/article/27/12/21-1631_article
Alpha and Beta variants became dominant in Finland in spring 2021 but diminished by summer. Outbreaks were mostly seeded by a few introductions. (Joshua may point out that this is not of interest because no club from Finland has reached the Champions league).
Bugembe DL, Phan MVT, Abias AG, et al. SARS-CoV-2 variants, South Sudan, January–March 2021. Emerg Infect Dis October 27, 2021.
Patterns of SARS-CoV-2 viral genomics in South Sudan in the second wave of infections during February–March 2021, showing circulation of B.1.525 (Eta) as well as the variant A.23.1. The B.1.525 genome encodes a deletion in the N-terminal domain (NTD) at spike positions 69 and 79, as well as being present in B.1.1.7 and many other global variants. A deletion in the spike NTD at positions 141–146 may help in evasion of host immune responses. (Same. South Sudan has presumably never even tried to reach Champions league).
31 October
Zhang J, Xiao T, Cai Y, et al. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science, October 26, 2021. https://www.science.org/doi/10.1126/science.abl9463
Delta spike can fuse membranes more efficiently at low levels of cellular receptor ACE2; also, its pseudotyped viruses infect target cells substantially faster than the other five variants, possibly accounting for its heightened transmissibility.
30 October
Singanayagam A, Hakki S, Dunning J, et al. Community transmission and viral load kinetics of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study. Lancet Inf Dis October 29, 2021. https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(21)00648-4/fulltext
Same. Vaccination is not sufficient to prevent Delta. The authors longitudinally followed index cases and their contacts (regardless of symptoms) in the community early after exposure to the Delta, performing daily quantitative RT-PCR for 14–20 days. Secondary attack rate in fully vaccinated household contacts was high at 25%, but this value was lower than that of unvaccinated contacts (38%). However, fully vaccinated individuals with breakthrough infections had peak viral load similar to unvaccinated cases.
29 October
Hitchings MD, Ranzani OT, Dorion M et al. Effectiveness of ChAdOx1 vaccine in older adults during SARS-CoV-2 Gamma variant circulation in São Paulo. Nat Commun October 27, 12, 6220 (2021). https://www.nature.com/search?q=COVID&order=date_desc
The AstraZeneca vaccine protects well from Gamma. During a period of high prevalence of the Gamma variant, effectiveness of the two-dose schedule was 77.9% (95% CI: 69.2–84.2) against COVID-19, 87.6% (95% CI: 78.2–92.9) against hospitalization, and 93.6% (95% CI: 81.9–97.7) against death.
28 October
Arora P, Rocha C, Kempf A. et al. The spike protein of SARS-CoV-2 variant A.30 is heavily mutated and evades vaccine-induced antibodies with high efficiency. Cell Mol Immunol October 25, 2021. https://www.nature.com/articles/s41423-021-00779-5
The variant A.30 (also termed A.VOI.V2), which was detected in several patients in Angola and Sweden in Spring 2021 and likely originated in Tanzania, can evade control by vaccine-induced antibodies and might show an increased capacity to enter cells in a cathepsin L-dependent manner, which may aid particularly in an extrapulmonary spread. Close monitoring warranted.
27 October
Taylor CA, Patel K, Pham H, et al. Severity of Disease Among Adults Hospitalized with Laboratory-Confirmed COVID-19 Before and During the Period of SARS-CoV-2 B.1.617.2 (Delta) Predominance — COVID-NET, 14 States, January–August 2021. MMWR Morb Mortal Wkly Rep. ePub: 22 October 2021. https://www.cdc.gov/mmwr/volumes/70/wr/mm7043e1.htm?s_cid=mm7043e1_w
Rates of COVID-19–associated hospitalization in adults increased during July–August 2021 as the Delta variant became predominant in the United States. Although Delta is more transmissible, this study did not find significantly higher proportions of hospitalizations with ICU admission, receipt of IMV, or in-hospital death in non-pregnant hospitalized adults.
25 October
Chagla Z, Ma H, Sander B, et al. Assessment of the Burden of SARS-CoV-2 Variants of Concern Among Essential Workers in the Greater Toronto Area, Canada. JAMA Netw Open October 19, 2021, 4(10):e2130284. doi:10.1001/jamanetworkopen.2021.30284
No surprise: VOC of SARS-CoV-2, similar to wildtype SARS-CoV-2, are disproportionately associated with neighborhoods with lower income and with a higher proportion of essential workers.
Abu-Raddad LJ, Chemaitelly H, Ayoub HH, et al. Severity, criticality, and fatality of the SARS-CoV-2 Beta variant Clinical Infectious Diseases, October 17, 2021 ciab909, https://doi.org/10.1093/cid/ciab909
The Beta (B.1.351) variant of COVID-19 disease was investigated in Qatar. Compared to the Alpha (B.1.1.7) variant, the odds of progressing to severe disease were 1.24-fold (95% CI: 1.11-1.39) higher for Beta. And the odds of COVID-19 death were 1.57-fold (95% CI: 1.03-2.43) higher for Beta.
21 October
Pouwels KB, Pritchard E, Matthews PC, et al. Effect of Delta variant on viral burden and vaccine effectiveness against new SARS-CoV-2 infections in the UK. Nat Med October 14, 2021. https://doi.org/10.1038/s41591-021-01548-7
Vaccination with two doses of BNT162b2 or ChAdOx1 substantially reduces the risk of new PCR-positive SARS-CoV-2 infections. However, while the two vaccines provided similar benefits when Alpha was dominant, benefits from two ChAdOx1 doses were reduced more in the presence of Delta than for two BNT162b2 doses, although two ChAdOx1 doses still provide similar protection as that from previous natural infection. With Delta, infections occurring after two vaccinations had similar peak viral burden as those in unvaccinated individuals. Vaccination reduces new infections, but effectiveness and attenuation of peak viral burden are reduced with Delta.
2 October
Choi A, Koch M, Wu K, et al. Serum Neutralizing Activity of mRNA-1273 Against SARS-CoV-2 Variants. J Virology 22 September 2021. https://journals.asm.org/doi/10.1128/JVI.01313-21
Minimal effects on neutralization titers against Alpha. Other VOCs such as Beta, Gamma and Delta, showed significantly decreased neutralization titers ranging from 2.1-fold to 8.4-fold reductions compared with D614G, although all remained susceptible to mRNA-1273–elicited serum neutralization.
22 September
Tao K, Tzou PL, Nouhin J, et al. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet September 19, 2021. https://www.nature.com/articles/s41576-021-00408-x
Brilliant review. And the best Figure of the day (Figure 1, your new bedroom poster)!
19 September
Blanquart F, Abad C, Ambroise J, et al. Characterisation of vaccine breakthrough infections of SARS-CoV-2 Delta and Alpha variants and within-host viral load dynamics in the community, France, June to July 2021. Euro Surveill September 15, 2021;26(37). https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2021.26.37.2100824
A French study, confirming other reports on Delta: higher viral load, longer duration of infection and, probably most important, similar values among fully vaccinated individuals and those who were not.
Luna-Muschi A, Borges IC, de Faria E, et al. Clinical features of COVID-19 by SARS-CoV-2 Gamma variant: a prospective cohort study of vaccinated and unvaccinated healthcare workers. J Infection September 16, 2021. https://www.journalofinfection.com/article/S0163-4453(21)00474-6/fulltext
Some differences in the clinical presentation between Gamma variant (P1 from Brazil) and non-VoC infection with a decreased frequency of hyposmia/anosmia and dysgeusia in Gamma variant cases.
15 September
Burugorri-Pierre C, Lafuente-Lafuente C, Oasi C, et al. Investigation of an Outbreak of COVID-19 in a French Nursing Home With Most Residents Vaccinated. JAMA Netw Open September 13, 2021; 4:e2125294. https://jamanetwork.com/journals/jamanetworkopen/fullarticle/2783985?resultClick=1
An outbreak in Biscarrosse, France, due to the B.1.1.7 variant, indicating that SARS-CoV-2 vaccination may not be sufficient as the sole means to prevent COVID-19 in nursing homes. Of 74 residents, 70 were fully and 2 were partially vacccinated. Among the 17 infected residents, one was unvaccinated, two were partially and were 14 fully vaccinated. Eight residents developed severe disease, 2 were hospitalized, and 1 individual (the unvaccinated resident) died.
12 September
Oude Munnink BB, Worp N, Nieuwenhuijse DF, et al. The next phase of SARS-CoV-2 surveillance: real-time molecular epidemiology. Nat Med September 9, 2021. https://www.nature.com/articles/s41591-021-01472-w
This Review summarizes the current knowledge on key viral mutations and variants.
10 September
Grint DJ, Wing K, Houlihan C, et al. Severity of SARS-CoV-2 alpha variant (B.1.1.7) in England. Clin Infect Dis September 6, 2021, ciab754, https://doi.org/10.1093/cid/ciab754
Using data from 185,234 people who tested positive for SARS-CoV-2 in the community, in fully adjusted analysis accounting for individual-level demographics and comorbidities as well as regional variation in infection incidence, the authors found alpha associated with 73% higher hazards of all-cause death and 62% higher hazards of hospital admission, compared to wild-type virus.
Tostanoski LH, Yu J, Mercado NB, et al. Immunity elicited by natural infection or Ad26.COV2.S vaccination protects hamsters against SARS-CoV-2 variants of concern. Science Translational Medicine September 7, 2021. https://www.science.org/doi/10.1126/scitranslmed.abj3789
See title. The Ad26.COV2.S vaccine (J&J) induced cross-reactive binding and neutralizing antibodies that were reduced against the B.1.351 strain (Beta) compared with wild type, but nevertheless still provided robust protection against B.1.351 challenge, as measured by weight loss and pathology scoring in the lungs.
8 September
Mlcochova P, Kemp S, Dhar MS, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature September 6, 2021. https://www.nature.com/articles/s41586-021-03944-y
This study combined in vitro experimentation and molecular epidemiology, confirming increased replication fitness and reduced sensitivity of SARS-CoV-2 B.1.617.2 (Delta) to neutralizing antibodies.
5 September
Strålin K, Bruce D, Wahlsttröm E, et al. Impact of the Alpha VOC on disease severity in SARS-CoV-2-positive adults in Sweden. J Infection August 30, 2021. https://www.journalofinfection.com/article/S0163-4453(21)00448-5/fulltext
Large study from Sweden. Alpha positive individuals had significantly higher rates of hospitalization (2.6% vs. 1.2%) and severe illness than variants of concern negative individuals overall, but the numbers were too small to evaluate differences in severity rate among hospitalized individuals.
31 August
Twohig KA, Nyberg T, Zaidi A, et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Inf Dis August 27, 2021. https://doi.org/10.1016/S1473-3099(21)00475-8
Large national study from England, indicating a higher hospital admission or emergency care attendance risk for patients with COVID-19 infected with delta compared with alpha. The HR of hospital admission within 14 days was 2.26 (95% CI 1.32–3.89) after stratification and regression adjustment for confounders.
Lam-Hine T, McCurdy SA, Santora L, et al. Outbreak Associated with SARS-CoV-2 B.1.617.2 (Delta) Variant in an Elementary School — Marin County, California, May–June 2021. MMWR 27 August 2021. https://www.cdc.gov/mmwr/volumes/70/wr/mm7035e2.htm?s_cid=mm7035e2_w
Delta is fast. An unvaccinated infected teacher continued to work for 2 days before receiving a test. On occasion during this time, the teacher read aloud unmasked to the class despite school requirements to mask while indoors. Results: 26 infections. Students were seated in five rows; despite masking, the attack rate in the two rows seated closest to the teacher’s desk was 80% (8/10) and was 28% (4/14) in the three back rows.
6 August
Kimura I, Kosugi Y, Wu J, et al. SARS-CoV-2 Lambda variant exhibits higher infectivity and immune resistance. bioRxiv 2021, posted 28 July. Full-text: https://doi.org/10.1101/2021.07.28.454085
More about the Lambda variant (also known as the C.37 lineage which is now spreading in South American countries such as Peru, Chile, Argentina, and Ecuador).
The authors describe three mutations or variants, the RSYLTPGD246-253N, L452Q and F490S mutations, which respectively confer resistance to vaccine-induced antiviral immunity. Additionally, the T76I and L452Q mutations contribute to enhanced viral infectivity.
18 July
Ferreira I, Kemp S, Datir R, et al. SARS-CoV-2 B.1.617 mutations L452 and E484Q are not synergistic for antibody evasion. J Infect Dis July 14, 2021, jiab368. https://academic.oup.com/jid/advance-article/doi/10.1093/infdis/jiab368/6321359
Spike bearing L452R and E484Q (as seen in Delta) confers modestly reduced sensitivity to BNT162b2 mRNA vaccine-elicited antibodies. The effect is similar in magnitude to the loss of sensitivity conferred by L452R or E484Q alone.
17 July
Williams SV, Vusirikala A, Ladhani SN, et al. An outbreak caused by the SARS-CoV-2 Delta (B.1.617.2) variant in a care home after partial vaccination with a single dose of the COVID-19 vaccine Vaxzevria, London, England, April 2021. Euro Surveill. 2021;26(27):pii=2100626. https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2021.26.27.2100626
The Delta attack rate after the first dose of Astra Zeneca’s vaccine was 35.7% (5/14) for staff and 81.3% (13/16) for residents, indicating lower protection in people who had received only one dose of the Vaxzevria vaccine within 3 months. Reassuringly, though, hospitalisation was uncommon and there were no deaths, providing some evidence that a single dose of vaccine may be protective against severe disease following infection with the Delta variant.
13 July
Souza WM, Amorim MR, Sesti-Costa R, et al. Neutralisation of SARS-CoV-2 lineage P.1 by antibodies elicited through natural SARS-CoV-2 infection or vaccination with an inactivated SARS-CoV-2 vaccine: an immunological study. Lancet Microbe July 08, 2021. https://www.thelancet.com/journals/lanmic/article/PIIS2666-5247(21)00129-4/fulltext
SARS-CoV-2 lineage P.1 (gamma) might escape neutralization by antibodies generated in response to polyclonal stimulation against previously circulating variants of SARS-CoV-2. Plasma from individuals previously infected with SARS-CoV-2 had an 8.6 times lower neutralizing capacity against the P.1 isolates.
12 July
Today a Delta special. Delta, also known as lineage B.1.617.2 (first seen in India), has some mutations in the spike protein (see below), which enhance transmissibility and decrease recognition of the immune system. In many countries, Delta has become the predominant variant. We’ll have encouraging data on effectiveness of several vaccines and of molnupiravir, as well as a pessimistic outbreak in a gym.
Chemaitelly H, Yassine HM, Benslimane FM, et al. mRNA-1273 COVID-19 vaccine effectiveness against the B.1.1.7 and B.1.351 variants and severe COVID-19 disease in Qatar. Nat Med July 7, 2021. https://www.nature.com/articles/s41591-021-01446-y
Real world effectiveness of Moderna’s vaccine against SARS-CoV-2 variants of concern in Qatar, using a matched case-control study design. Effectiveness against B.1.1.7 infection (alpha) was 88% ≥ 14 days after the first dose but before the second dose and was 100% ≥ 14 days after the second dose. Analogous effectiveness against B.1.351 infection (beta) was 61% after the first dose and 96% after the second dose. Effectiveness against any severe, critical or fatal COVID-19 disease due to SARS-CoV-2 infection was 82% and 96% after the first and second dose, respectively. Compared to beta (64%) and alpha (10%), the percentage of delta was low (3%). However, it is encouraging to see the high effectiveness against beta which constitutes the variant of the greatest concern in regard to immune escape (much more so than delta).
Lazarevic I, Pravica V, Miljanovic D, et al. Evasion of SARS-CoV-2 Emerging Variants: What Have We Learnt So Far? Viruses June 22, 2021,13, 1192. https://www.mdpi.com/1999-4915/13/7/1192
Nice review on variants of concern, summarizing current knowledge. A must read.
Yadav PD, Sapkal GN, Ella R, et al. Neutralization of Beta and Delta variant with sera of COVID-19 recovered cases and vaccinees of inactivated COVID-19 vaccine BBV152/Covaxin. Journal of Travel Medicine Jul 6, 2021, taab104, https://academic.oup.com/jtm/advance-article/doi/10.1093/jtm/taab104/6316247?searchresult=1
An inactivated SARS-CoV-2 vaccine, BBV152, was rolled out under the national COVID-19 vaccination program in India. BBV152 was found to confer significant protection against delta (and beta).
Planas D, Veyer D, Baidaliuk A. et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature July 8, 2021. https://www.nature.com/articles/s41586-021-03777-9
Delta partially but significantly escapes neutralizing mAbs, and polyclonal antibodies elicited by previous SARS-CoV-2 infection or vaccination. Sera from individuals having received one dose of either the Pfizer or AstraZeneca vaccines barely inhibited the delta variant. Administration of two doses generated a neutralizing response in 95% of individuals, with titers 3-to-5-fold lower against delta than alpha.
Abdelnabi R, Foo CS, De Jonghe S, et al. Molnupiravir inhibits the replication of the emerging SARS-CoV-2 variants of concern (VoCs) in a hamster infection model. The Journal of Infectious Diseases Juily 9, 2021, jiab361, https://academic.oup.com/jid/advance-article/doi/10.1093/infdis/jiab361/6318434
Molnupiravir, currently in Phase II clinical trials, has worked well in hamsters infected with either Wuhan strain, with the delta or alpha variants.
Dougherty K, Mannell M, Naqvi O, Matson D, Stone J. SARS-CoV-2 B.1.617.2 (Delta) Variant COVID-19 Outbreak Associated with a Gymnastics Facility — Oklahoma, April–May 2021. MMWR Morb Mortal Wkly Rep. ePub: 9 July 2021. https://www.cdc.gov/mmwr/volumes/70/wr/mm7028e2.htm?s_cid=mm7028e2_w
This delta outbreak may indicate that return to gym in 2021 is probably not the best idea. During April 15–May 3, 2021, 47 COVID-19 cases (all 21 tested cases were delta) were linked to a gymnastics facility. The overall facility and household attack rates were 20% and 53%, respectively. Of note, four persons (9%) had received 2 doses of either the Moderna or Pfizer-BioNTech or a single dose of the Johnson & Johnson vaccine ≥ 14 days before a positive test result. Several potential risk factors for transmission were identified, including not using masks among active participants, coupled with increased respiration during active sport participation (further, facility policy was that all persons not actively participating wear masks, but this policy was not always followed); poor facility ventilation; staff members training multiple cohorts, etc.
10 July
Tarke A, Sidney J, Methot N, et al. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep July 01, 2021. https://www.cell.com/cell-reports-medicine/fulltext/S2666-3791(21)00204-4
The SARS-CoV-2 variants B.1.1.7, B.1.351, P.1 and CAL.20C do not significantly disrupt the total SARS-CoV-2 T cell reactivity. T cells of exposed donors or vaccinees effectively recognized SARS-CoV-2 variants.
10 July
Tarke A, Sidney J, Methot N, et al. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep July 01, 2021. https://www.cell.com/cell-reports-medicine/fulltext/S2666-3791(21)00204-4
The SARS-CoV-2 variants B.1.1.7, B.1.351, P.1 and CAL.20C do not significantly disrupt the total SARS-CoV-2 T cell reactivity. T cells of exposed donors or vaccinees effectively recognized SARS-CoV-2 variants.
8 July
Widera M, Wilhelm A, Hoehl S, et al. Limited neutralization of authentic SARS-CoV-2 variants carrying E484K in vitro. The Journal of Infectious Diseases 05 July 2021, jiab355, https://academic.oup.com/jid/advance-article/doi/10.1093/infdis/jiab355/6314691
This in vitro study using authentic SARS-CoV-2 confirms that, in contrast to vaccine-elicited sera, bamlanivimab and casirivimab may not provide efficacy against SARS-CoV-2 variants B.1.351 (beta) and P.2 (zeta), both harboring the E484K substitution. Since imdevimab was able to efficiently neutralize both variants, therapeutical treatment with the REGN-COV-2 combination remains effective.
7 July
Lustig Y, Zuckerman N, Nemet I, et al. Neutralising capacity against Delta (B.1.617.2) and other variants of concern following Comirnaty (BNT162b2, BioNTech/Pfizer) vaccination in health care workers, Israel. Euro Surveill Jul 1, 2021;26(26):pii=2100557. https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2021.26.26.2100557
Using micro-neutralization assays with sera obtained after Comirnaty (BNT162b2, BioNTech/Pfizer) vaccination in 36 healthcare workers, the authors show that, despite somewhat reduced neutralization capacity, mRNA vaccination induces a substantial antibody response for the Delta VOC as well.
5 July
McCallum M, Bassi J, De Marco A, et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science 01 Jul 2021: eabi7994. https://science.sciencemag.org/content/early/2021/06/30/science.abi7994
How the B.1.427/B.1.429 variant (“California”) evades immune responses. The L452R mutation reduced neutralizing activity of 14 out of 34 RBD-specific monoclonal antibodies (mAbs). The S13I and W152C mutations resulted in total loss of neutralization for 10 out of 10 NTD-specific mAbs. In this case, the NTD antigenic supersite was remodeled by a shift of the signal peptide cleavage site and formation of a new disulfide bond, as revealed by mass spectrometry and structural studies.
2 July
Hoffmann M, Hofmann-Winkler H, Krüger N, et al. SARS-CoV-2 variant B.1.617 is resistant to Bamlanivimab and evades antibodies induced by infection and vaccination. Cell Reports June 28, 2021. https://www.cell.com/cell-reports/fulltext/S2211-1247(21)00828-7
The spike protein of B.1.617 (delta) harbors two mutations in the receptor-binding domain, which interacts with the ACE2 receptor and constitutes the main target of neutralizing antibodies. The B.1.617 spike protein can facilitate entry into lung cells with slightly increased efficiency and shows that entry can be blocked by soluble ACE2 and camostat. B.1.617 also evades antibodies induced by infection or vaccination, although less so than the B.1.351 variant.
30 June
Pokhrel S, Kraemer BR, Burkholz S. et al. Natural variants in SARS-CoV-2 Spike protein pinpoint structural and functional hotspots with implications for prophylaxis and therapeutic strategies. Sci Rep June 23, 2021, 11, 13120. https://www.nature.com/articles/s41598-021-92641-x
This analysis of the frequency of variants throughout the S protein of SARS-CoV-2 identified regions of high and low divergence, which may aid in developing effective prophylactic and therapeutic treatments.
29 June
Gobeil MC, Janowska K, McDowell S, et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science 24 Jun 2021: eabi6226. https://science.sciencemag.org/content/early/2021/06/23/science.abi6226?rss=1
This study reveals allosteric effects of mutations and mechanistic differences that drive either inter-species transmission or escape from antibody neutralization. In other words, this study explains the rapid spread of variants. There is an increased binding to ACE2, mediated both by affinity enhancing substitutions in the RBD and increased propensity for the receptor-accessible RBD up states. All variants showed increased ACE2 receptor binding and increased propensity for RBD up states. A local destabilizing effect of the RBD E484K mutation was implicated in resistance of the B.1.1.28/P.1 (Brazil) and B.1.351 (South Africa) variants to neutralizing antibodies.
27 June
Today we focus on two issues: variants of concern and long COVID. First, read about some new variants of concern (VOC) and how SARS-CoV-2 has evolved to enhance viral fitness and immune evasion (and that the worst is yet to come!). Second, we summarize some long COVID studies in non-hospitalized cohorts, long-term data and studies addressing different issues such as anosmia or tauopathy.
25 June
Krause PR, Fleming TR, Longini IM, et al. SARS-CoV-2 Variants and Vaccines. NEJM June 23, 2021. https://www.nejm.org/doi/full/10.1056/NEJMsr2105280?query=featured_home
Special report on variants. So far, there is no good evidence that currently identified variants of concern evade the most important vaccine effect — that of prevention of severe disease.
Bager P, Wohlfahrt J, Fonager J, et al. Risk of hospitalisation associated with infection with SARS-CoV-2 lineage B.1.1.7 in Denmark: an observational cohort study. Lancet June 22, 2021. https://doi.org/10.1016/S1473-3099(21)00290-5
In this large study on 50,958 individuals, however, B.1.1.7 was associated with an adjusted RR of 1.42 (95% CI 1.25–1.60; p<0.0001). The adjusted RR was increased in all strata of age and calendar period—the two covariates with the largest contribution to confounding of the crude RR.
20 June
Campbell F, Archer B, Laurenson-Schafer H, et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Eurosurveillance June 2021; 26(24):pii=2100509. https://doi.org/10.2807/1560-7917.ES.2021.26.24.2100509
This analysis of the effective reproduction number and global spread of SARS-CoV-2 variants with data available by 3 June 2021 suggest that B.1.617.2 is expected to rapidly out-compete other variants and become the dominant circulating lineage over the coming months.
Jacobson KB, Pinsky BA, Rath ME, et al. Post-vaccination SARS-CoV-2 infections and incidence of presumptive B.1.427/B.1.429 variant among healthcare personnel at a northern California academic medical center. Clin Inf Dis June 17, 2021, ciab554, https://doi.org/10.1093/cid/ciab554
Presumptive B.1.427/B.1.429 was not more prevalent in post-vaccine cases than in unvaccinated SARS-CoV-2 healthcare personnel.
19 June
Motozono C, Toyoda M, Zahradnik J, et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe June 14, 2021. https://doi.org/10.1016/j.chom.2021.06.006
This study demonstrates that at least two naturally occurring mutations in the SARS-CoV-2 RBM, L452R and Y453F, escape HLA restricted cellular immunity and further promote affinity toward the viral receptor ACE2. The L452R mutation also increases the stability of the S protein and viral infectivity.
Liu C, Ginn HM, Dehnirattisai W, et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell June 16, 2021. https://doi.org/10.1016/j.cell.2021.06.020
The ability of monoclonal antibodies, convalescent and vaccine sera to neutralize B.1.617.1 and B.1.617.2 is reduced when compared with ancestral Wuhan strains but there is no evidence of widespread antibody escape as seen with B.1.351. However, B.1.351 and P.1 sera showed markedly more reduction in neutralization of B.1.617.2 suggesting that individuals previously infected by these variants may be more susceptible to re-infection by B.1.617.2.
17 June
Nyberg TR, Twohig KA, Harris RJ, et al. Risk of hospital admission for patients with SARS-CoV-2 variant B.1.1.7: cohort analysis. BMJ 2021 June 15, 2021; 373. https://www.bmj.com/content/373/bmj.n1412
This retrospective analysis identified through community testing in England indicated that the risk of hospital admission within 14 days after a positive test was 1.52 (1.47 to 1.57) times higher for patients infected with the B.1.1.7 variant compared with those infected with wild-type variants, after adjustment for age, sex, deprivation, ethnicity, region, and week of diagnosis.
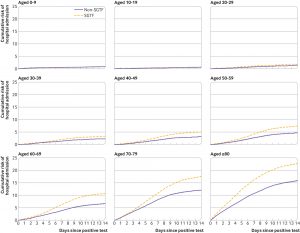
Cumulative risk of hospital admission within 1-14 days after positive SARS-CoV-2 test, by age group. Risks were estimated with Cox regression stratified by S-gene target failure (SGTF) status and age group, adjusted for sex, index of multiple deprivation fifth, ethnicity, region of residence, and calendar week (potential confounders set to mean covariate levels)
15 June
Buchan SA, Tibebu S, Daneman N, et al. Increased household secondary attacks rates with Variant of Concern SARS-CoV-2 index cases. Clin Inf Dis June 9, 2021, ciab496. https://academic.oup.com/cid/advance-article/doi/10.1093/cid/ciab496/6295404
Huge study on 5617 index cases and 3397 secondary cases. In a propensity score matched analysis, the secondary attack rate for variant of concern (VOC) index cases was 1.31 times higher than non-VOC index cases. This increase was particularly accentuated for asymptomatic or pre-symptomatic index cases.
13 June
Liu J, Liu Y, Xia H, et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature June 10, 2021. https://www.nature.com/articles/s41586-021-03693-y
Good news. A total of 20 human sera, drawn 2 or 4 weeks after two doses of BNT162b2, neutralized engineered SARS-CoV-2 with different spike proteins of the delta variants (from India). B.1.617.1 was the least neutralized, probably due to the presence of both L452R and E484Q substitutions at the receptor binding site. Nevertheless, all variants were neutralized by all tested sera at titers ≥ 40.
Alter G, Yu J, Liu J, et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature June 9, 2021. https://www.nature.com/articles/s41586-021-03681-2
Median pseudovirus neutralizing antibody titers induced by Ad26.COV2.S (Johnson & Johnson) were 5.0-fold lower against the B.1.351 variant and 3.3-fold lower against the P.1 variant as compared with the original Wuhan strain, which is comparable to other vaccines. Functional non-neutralizing antibody responses and CD8+ and CD4+ T cell responses were largely preserved.
10 June
Wang R, Zhang W, Ge J, et al. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity Jun 8, 2021. https://www.cell.com/immunity/fulltext/S1074-7613(21)00247-8
The South African variant beta (B.1.351) was the most resistant to current monoclonal antibodies and convalescent plasma, followed by the Brazilian variant gamma (P.1) and the UK variant alpha (B.1.1.7). This resistance hierarchy corresponded with deletions in the N-terminal domain and K417N/T, E484K and N501Y mutations in the receptor binding domain (RBD) of SARS-CoV-2.
8 June
Vanker A, McGeer A, O’Byrne G, et al. Adverse Outcomes Associated with SARS-CoV-2 variant B.1.351 Infection in Vaccinated Residents of a Long Term Care Home, Ontario, Canada. Clin Inf Dis, June 6, 2021, ciab523, https://academic.oup.com/cid/advance-article/doi/10.1093/cid/ciab523/6294072
Another outbreak of B.1.351 (south Africa) in a long term care home (LTCH) in Canada. Of the 9 residents (vaccinated with B162.B2 from BionTech/Pfizer) and infected with B.1.351, 4 developed hypoxemia. Of these, 3 (including both hospitalized patients) died.
7 June
Lesho E, Corey B, Lebreton F, et al. Emergence of the E484K Mutation in SARS-CoV-2 Lineage B.1.1.345 in Upstate New York. Clin Infect Dis June 4, 2021, ciab507, https://academic.oup.com/cid/advance-article/doi/10.1093/cid/ciab507/6292251
Case series (n=15) of infections with a SARS-CoV-2 B.1.1.345 variant (from New York) carrying the E484K mutation. E484K probably has a role in antibody evasion.
3 June
Callaway E. Coronavirus variants get Greek names — but will scientists use them? Nature NEWS June 1, 2021. https://www.nature.com/articles/d41586-021-01483-0
From Alpha to Omega, the new labelling system aims to avoid confusion and stigmatization. Ewen Callaway explains why the new system could be useful.
31 May
Marot S, Malet I, Leducq V, et al. Neutralization heterogeneity of United Kingdom and South-African SARS-CoV-2 variants in BNT162b2-vaccinated or convalescent COVID-19 healthcare workers. Clinical Infectious Diseases, May 29, 2021, ciab492, https://academic.oup.com/cid/advance-article/doi/10.1093/cid/ciab492/6288468
Two findings from this small study, assessing the neutralizing activity of sera from 15 convalescent COVID-19 HCW and from 29 BNT162b2 vaccinated HCW against B.1.1.7 (UK) and B.1.351 (South Africa): 1. One shot is not enough, questioning the timing of the dosing interval of BNT162b2. After two shots, however, most participants displayed a neutralizing activity ≥ 1:10 which could be at least indicative of a potential protection against variants.
30 May
Singh J, Rahman SA, Ehtesham NZ, et al. SARS-CoV-2 variants of concern are emerging in India. Nat Med May 27, 2021. https://doi.org/10.1038/s41591-021-01397-4
The sudden surge in COVID-19 cases in India coincides with high prevalence of more transmissible variants, associated with diagnostic test failures and antibody escape. This paper gives an overview.
26 May
Ratckliff J, Nguyen D, Fish M, et al. Virological and serological characterization of critically ill patients with COVID-19 in the UK: Interactions of viral load, antibody status and B.1.1.7 variant infection. J Inf Dis 24 May 2021, jiab283, https://academic.oup.com/jid/advance-article/doi/10.1093/infdis/jiab283/6283590
Among 1274 subjects, viral loads were higher in B.1.1.7 infected individuals than those infected with wild-type, but only in those who had already seroconverted for anti-SARS-CoV-2 antibody, indicating less effective clearance by innate and adaptive immune responses.
22 May
Yuan M, Huang D, Lee CC, et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science 20 May 2021:eabh1139. https://science.sciencemag.org/content/early/2021/05/19/science.abh1139
The B.1.1.7 lineage has acquired N501Y, the B.1.351 and P.1 lineages share this mutation plus K417N/T and E484K, and the “California” variants have an L452R mutation that is also present in the Indian variant B.1.617 with E484Q. Binding and neutralization of the two most frequently elicited antibody families are shown to be abrogated by K417N, E484K, or both. These effects can be structurally explained by their extensive interactions with receptor-binding site nAbs. Since nAbs to the more conserved, cross-neutralizing CR3022 and S309 sites were largely unaffected, these sites are promising targets to avoid interference by SARS-CoV-2 mutations observed to date.
21 May
Meister TL, Fortmann J, Todt D, et al. Comparable environmental stability and disinfection profiles of the currently circulating SARS-CoV-2 variants of concern B.1.1.7 and B.1.351. J Inf Dis 2021, May 16, 2021. https://academic.oup.com/jid/advance-article/doi/10.1093/infdis/jiab260/6276396
Treatment with heat, soap and ethanol revealed similar inactivation profiles indicative of a comparable susceptibility to disinfection. Surface stability was comparable on steel, silver, copper and face masks.
12 May
Lee LY Rozmanowski S, Pang M, et al. SARS-CoV-2 infectivity by viral load, S gene variants and demographic factors and the utility of lateral flow devices to prevent transmission. Clinical Infectious Diseases May 11, 2021, ciab421, https://doi.org/10.1093/cid/ciab421
This large-scale analysis of combined SARS-CoV-2 contact tracing and testing data from England involving > 2 million contacts of PCR-confirmed cases shows that SARS-CoV-2 infectivity is associated with index case viral load, including after adjustment for demographic factors and type of contact event. SGTF (a proxy for the B.1.1.7 variant), increased transmission by ~ 50% at most viral loads. Except SGTF, there was no significant interaction between Ct values and any other variables in the analysis. Thus, the effect of viral load on infectivity is generalizable across populations and settings.
11 May
Staub T, Arendt V, de la Vega EC. Case series of four re-infections with a SARS-CoV-2 B.1.351 variant, Luxembourg, February 2021. Eurosurveillance May 6, 2021. https://www.eurosurveillance.org/content/eurosurveillance/26/18
Four female HCW from the same hospital. All recovered and their re-infections were not severe.
29 April
Resende PC, Bezerra JF, Vasconcelos RHT, Arantes I, Appolinario L, Mendonça AC, et al. Severe acute respiratory syndrome coronavirus 2 P.2 lineage associated with reinfection case, Brazil, June–October 2020. Emerg Infect Dis April 21, 2021 Jul [date cited] https://wwwnc.cdc.gov/eid/article/27/7/21-0401_article
Case report of a 37-year-old healthcare worker from the northeastern region of Brazil who experienced two clinical episodes, 116 days apart. The two infections were caused by the most prevalent lineage in Brazil, B.1.1.33, and the emerging lineage P.2.
26 April
Funk T, Pharris A, Spiteri G, et al. Characteristics of SARS-CoV-2 variants of concern B.1.1.7, B.1.351 or P.1: data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Euro Surveill 2021;26(16):pii=2100348. https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2021.26.16.2100348
This analysis outlines the characteristics of SARS-CoV-2 variants of concern (VOC) infections in seven EU/EEA countries, including 19,995 VOC and 3348 non-VOC cases, suggesting a higher risk for hospitalization, and also for ICU admission in age groups < 60 years for B.1.1.7, B.1.351 and P.1. The matched multivariable analysis found that VOC had significantly higher odds of hospitalization than non-VOC cases (aOR: 1.6–4.2).
22 April
Munitz A, Yechezkel M, Dickstein Y. BNT162b2 Vaccination Effectively Prevents the Rapid Rise of SARS-CoV-2 Variant B.1.1.7 in high risk populations in Israel. Cell Rep Med April 17, 2021. https://www.cell.com/cell-reports-medicine/fulltext/S2666-3791(21)00080-X
Good news. Although B.1.1.7 is 45% more transmissible than the wild type strain, becoming the dominant lineage in Israel within a few weeks, focused RT-PCR testing and prioritized vaccination programs can prevent the spread of the B.1.1.7 variant in the elderly.
Deng X, Garcia-Knight MA, Khalid MM, et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell April 20, 2021. https://www.cell.com/cell/fulltext/S0092-8674(21)00505-5
The “California” variant named B.1.427/B.1.429 to denote its 2 lineages, with 3 mutations, including L452R, emerged in May 2020 and increased from 0% to > 50% of sequenced cases from September 2020 to January 2021, showing a 19-24% increased transmissibility. Antibody neutralization assays revealed modest decreases (4.0 to 6.7-fold and 2.0-fold) in neutralizing titers from convalescent patients and vaccine recipients, respectively.
16 April
Faria NR, Mellan TA, Whittaker C, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 14 Apr 2021: eabh2644. https://science.sciencemag.org/content/early/2021/04/13/science.abh2644
The second wave of infection in Manaus was associated with the emergence and rapid spread of lineage P.1. that occurred mid-November 2020 and had acquired 17 mutations, including a trio in the spike protein (K417T, E484K and N501Y) associated with increased binding to the human ACE2 receptor. P.1 may be 1.7–2.4-fold more transmissible, and previous (non-P.1) infection provides 54–79% of the protection against infection with P.1 that it provides against non-P.1 lineages.
15 April
Graham MS, Sudre CH, May A, et al. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: an ecological study. Lancet Public Health April 12, 2021. https://www.thelancet.com/journals/lanpub/article/PIIS2468-2667(21)00055-4/fulltext
From Sept 28 to Dec 27, 2020, positive COVID-19 tests were reported by 36,920 COVID Symptom Study app users whose region was known and who reported as healthy on app sign-up. There were no changes in reported symptoms or disease duration associated with B.1.1.7.
Betton M, Livrozet M, Planas D, et al. Sera neutralizing activities against SARS-CoV-2 and multiple variants six month after hospitalization for COVID-19. Clinical Infectious Diseases 14 April 2020, ciab308, https://academic.oup.com/cid/advance-article/doi/10.1093/cid/ciab308/6225251?searchresult=1
This prospective study analyzed sera of 107 patients hospitalized with COVID-19 at 3 and 6 months post-infection. Decrease of IgG rates and serological assays becoming negative did not imply loss of neutralizing capacity. Sera collected at 6 months showed efficient neutralizing effects against the D614G, B.1.1.7 and P.1 variants but a significantly weaker activity against the B.1.351 variant.
14 April
Frampton D, Rampling R, Cross A, et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Inf Dis April 12, 2021. https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(21)00170-5/fulltext
It will be interesting to see whether this elegant study will gain as much media attraction as the studies showing the opposite: comparing 198 patients with B.1.1.7 infection and 143 with non-B.1.1.7 infection, the authors found NO evidence of an association between severe disease and death and lineage in unadjusted analyses or in analyses adjusted for hospital, sex, age, co-morbidities, and ethnicity. Viral load by proxy was higher in B.1.1.7 samples than in non-B.1.1.7 samples, as measured by cycle threshold value.
Ong SW, Young BE, Lye DC. Lack of detail in population-level data impedes analysis of SARS-CoV-2 variants of concern and clinical outcomes. Lancet Inf Dis April 12, 2021. https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(21)00201-2/fulltext
Comment on the previous study, followed by a nice overview. What do we know about the effects of B.1.1.7 on disease severity? Not enough. Confounding factors including health care resource use, demographic changes, and socio-behavioral trends affect clinical outcomes, including mortality, and are difficult to adjust for without detailed, robust, patient-level data. According to the authors, “careful epidemiologic and clinical assessment, coupled with a healthy skepticism, is important when assessing claims of the effect of these variants”. Agreed.
12 April
Shen X, Tang H, Pajon R, et al. Neutralization of SARS-CoV-2 Variants B.1.429 and B.1.351. NEJM April 7, 2021. https://www.nejm.org/doi/full/10.1056/NEJMc2103740?query=featured_home
Vaccine-elicited neutralizing antibodies are likely to remain effective against the B.1.429 variant (“California”). The modestly lower value in neutralization titers was similar to B.1.1.7, using serum from recipients of the mRNA-1273 (Moderna) and NVX-CoV2373 (Novavax). The magnitude of resistance seen with the B.1.351 variant is of greater concern. However, this is good news, because immune escape seems to be limited in most variants.
8 April
Wang GL, Wang ZY, Duan LJ, et al. Susceptibility of Circulating SARS-CoV-2 Variants to Neutralization. NEJM April 6, 2021. https://www.nejm.org/doi/full/10.1056/NEJMc2103022?query=featured_coronavirus
B.1.1.7 showed little resistance to the neutralizing activity of convalescent or vaccinee serum (inactivated-virus vaccines from Sinopharm and Sinovac were used), whereas B.1.351 showed more resistance to the neutralization of both convalescent serum (by a factor of 2) and vaccinee serum (by a factor of 2.5 – 3.3) than to the wild-type virus. Results are in line with previous studies with mRNA vaccines.
De Oliveira T, Lutucuta S, Nkengasong J, et al. A novel variant of interest of SARS-CoV-2 with multiple spike mutations is identified from travel surveillance in Africa. https://www.medrxiv.org/content/10.1101/2021.03.30.21254323v1
Not peer reviewed: a new variant was found in Angola in three travelers arriving from Tanzania who were tested together at the airport in mid-February. The variant, named A.VOI.V2, has 31 amino acid substitutions (11 in spike) and three deletions (all in spike). According to the authors, this warrants urgent investigation as the source country has a largely undocumented epidemic and few public health measures in place to prevent spread either within or outside of the country.
7 April
Hoffmann M, Zhang L, Krüger N, et al. SARS-CoV-2 mutations acquired in mink reduce antibody-mediated neutralization. Cell Report, April 2, 2021. 109017. https://www.cell.com/cell-reports/fulltext/S2211-1247(21)00331-4
Mutations frequently found in the S proteins of SARS-CoV-2 from mink were mostly compatible with efficient entry into human cells and its inhibition by soluble ACE2. In contrast, mutation Y453F reduced neutralization by casirivimab and by sera/plasma from COVID-19 patients. Infection of mink and other animal species should be prevented and it should be continuously monitored whether SARS-CoV-2 amplification in other wild or domestic animals occurs and changes critical biological properties of the virus.
6 April
Schuit M, Biryukov J, Beck K, et al. The stability of an isolate of the SARS-CoV-2 B.1.1.7 lineage in aerosols is similar to three earlier isolates. J Inf Dis April 2, 2021, jiab171, https://academic.oup.com/jid/advance-article/doi/10.1093/infdis/jiab171/6209391?searchresult=1
The stability of SARS-CoV-2 in aerosols does not vary greatly among the currently circulating lineages, including B.1.1.7, suggesting that the increased transmissibility associated with recent SARS-CoV-2 lineages is not due to enhanced survival in the environment.
Starr TN, Greaney AJ, Dingens AS, et al. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep Med April 01, 2021. https://www.cell.com/cell-reports-medicine/fulltext/S2666-3791(21)00071-9
Future efforts should diversify the epitopes targeted by antibodies to make them more resilient to antigenic evolution. Individual mutations that escape binding by bamlanivimab (BAM) and etesevimab (ESV) are combined in the B.1.351 and P.1 lineages (E484K escapes BAM, K417N/T escapes ESV). Additionally, the L452R mutation in the B.1.429 lineage escapes BAM. The authors also identified single amino acid changes that escape the combined BAM/ESV cocktail.
5 April
The SARS-CoV-2 variant with lineage B.1.351 clusters investigation team. Linked transmission chains of imported SARS-CoV-2 variant B.1.351 across mainland France, January 2021. Euro Surveill 2021;26(13):pii=2100333. https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2021.26.13.2100333
Two cases had travelled in mid-December 2020 with a group to Mozambique where they participated in a religious gathering and returned with the B.1.351 variant. A joint team of epidemiologists, public health workers and clinical and virological specialists co-operated across France to urgently investigate and initiate control measures. A total of 36 cases were analyzed. Believe it or not: “Another challenge was that some members of the clusters did not agree to answer questions”.
4 April
Graham C, Seow J, Huettner I, et al. Neutralization potency of monoclonal antibodies recognizing dominant and subdominant epitopes on SARS-CoV-2 Spike is impacted by the B.1.1.7 variant. Immunity April 01, 2021. https://www.cell.com/immunity/fulltext/S1074-7613(21)00135-7
To understand how mutations affect Spike antigenicity, the authors isolated and characterized > 100 monoclonal antibodies targeting epitopes on SARS-CoV-2 spike receptor-binding domain (RBD), N-terminal domain (NTD) and S2 from SARS-CoV-2-infected individuals. Mutations present in B.1.1.7 spike frequently conferred neutralization resistance to NTD-specific antibodies. Neutralization by RBD-specific nAbs remained largely unchanged.
3 April
Peng J, Liu J, Mann SA, et al. Estimation of secondary household attack rates for emergent spike L452R SARS-CoV-2 variants detected by genomic surveillance at a community-based testing site in San Francisco. Clinical Infectious Diseases 31 March 2021, ciab283, https://academic.oup.com/cid/advance-article/doi/10.1093/cid/ciab283/6206738?searchresult=1
The new lineages B.1.427 and B.1.429 (the “California” or “West Coast” variants) share S gene non-synonymous mutations at sites 13, 152, 452, and 614 and were seen during the December 2020 to February 2021 period when California was experiencing a huge peak. Whereas no instances of B.1.1.7, or independent N501Y mutations were detected in the sample population, the authors found a modest transmissibility increase of the West Coast variants. Household contacts exposed to these variants were at higher risk of infection compared to those exposed to lineages lacking these variants (0.36 vs 0.29, RR = 1.28; 95% CI: 1.00-1.64). Ct values did not differ.
2 April
Dejnirattisai W, Zhou D, Supasa P, et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell March 30, 2021. https://www.cell.com/cell/fulltext/S0092-8674(21)00428-1
All new strains (P.1 from Brazil, B.1.351 from South Africa and B.1.1.7 from the UK) have mutations in the ACE2 binding site with P.1 and B.1.351 having a virtually identical triplet: E484K, K417N/T and N501Y, conferring similar increased affinity for ACE2. Surprisingly, P.1 was significantly less resistant to naturally acquired or vaccine induced antibody responses than B.1.351, suggesting that changes outside the receptor-binding domain impact neutralization.
31 March
Francisco MA, Zavascki AP, Lamb WP, et al. Detection of SARS-CoV-2 lineage P.1 in patients from a region with exponentially increasing hospitalisation rate, February 2021, Rio Grande do Sul, Southern Brazil. Euro Surveill Accepted: 25 Mar 2021, 2021. Full text: https://doi.org/10.2807/1560-7917.ES.2021.26.12.2100276
From epidemiological week 6, starting on 7 February 2021, until 6 March, the number of hospitalizations for COVID-19 in Rio Grande do Sul, the southernmost state of Brazil in the South region, increased from 1738 inpatients to 6995 (3.8-fold). This resulted in the collapse of the state healthcare system. The overwhelming increase in hospitalizations temporally coincided with the finding that lineage P.1 became predominant (although a small number of specimens was taken).
28 March
Abbasi J. How the US Failed to Prioritize SARS-CoV-2 Variant Surveillance. JAMA. 2021 Mar 24. PubMed: https://pubmed.gov/33760030. Full-text: https://doi.org/10.1001/jama.2021.3368
Until late last fall, public health departments had no federal mandate or additional funding to sequence samples, although new variants were an inevitability.
27 March
McCormick KD, Jacobs JL, Mellors JW. The emerging plasticity of SARS-CoV-2. Science 2021, published 26 March. Full text: https://doi.org/10.1126/science.abg4493
Another excellent summary and extension of our Variants chapter. By John Mellors and colleagues.
26 March
Volz E, Mishra S, Chand M, et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature (2021). Full text: https://doi.org/10.1038/s41586-021-03470-x
The preprint we presented on 13 January now published in Nature: the authors describe the new SARS-CoV-2 lineage B.1.1.7 (AKA VOC 202012/01) which originated in England, late Summer to early Autumn 2020. The data indicate a transient shift in the age composition of reported cases, with a larger share of under-20-year-olds among B.1.1.7 cases than among historical cases. B.1.1.7 has a substantial transmission advantage with a 50% to 100% higher reproduction number.
25 March
Edara VV, Norwood C, Floyd K, et al. Infection and vaccine-induced antibody binding and neutralization of the B.1.351 SARS-CoV-2 variant. Cell Host Microbe 2021, published 20 March. Full text: https://doi.org/10.1016/j.chom.2021.03.009
Despite reduced antibody titers against the B.1.351 variant (first detected in South Africa), sera from infected and vaccinated individuals containing polyclonal antibodies to the spike protein could still neutralize SARS-CoV-2 B.1.351. The authors conclude that protective humoral immunity may be retained against this variant.
23 March
Hoffmann M, Arora P, Groß R, et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021, accepted 16 March. Full-text: https://doi.org/10.1016/j.cell.2021.03.036
The authors show that entry of all variants into human cells is susceptible to blockade by the entry inhibitors soluble ACE2, Camostat, EK-1 and EK-1-C4. In contrast, entry of B.1.351 and P.1 was partially (casirivimab) or fully (bamlanivimab) resistant to monoclonal antibodies. Moreover, entry of these variants was less efficiently inhibited by plasma from convalescent COVID-19 patients and sera from individuals vaccinated with the Pfizer-BioNTech vaccine.
20 March
Grint DJ, Wing K, Williamson E, et al. Case fatality risk of the SARS-CoV-2 variant of concern B.1.1.7 in England, 16 November to 5 February. Volume 26, Issue 11, 18 March 2021. https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2021.26.11.2100256
Evidence grows that B.1.1.7 is more dangerous. In this data drawn from the OpenSAFELY electronic health records secure research platform (covering 40% of England’s population registered with a general practitioner), there was a consistently higher (about two thirds) absolute risk of death by 28 days after a SARS-CoV-2-positive test in all groups stratified by age, sex and presence of co-morbidities.
Graphic
19 March
Paper of the Day
Wu K, Werner AP, Koch M. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine. NEJM March 17, 2021. https://www.nejm.org/doi/full/10.1056/NEJMc2102179
What about Moderna’s mRNA vaccine and the new variants? This study saw a decrease in titers of neutralizing antibodies against the P.1 variant, the B.1.427/B.1.429 variant, the B.1.1.7+E484K variant, and the B.1.351 variant as well as a subset of its mutations in the RBD. Protection against these lineages remains “to be determined”.
17 March
Challen R, Brooks-Pollock E, Read JM, Dyson L, Tsaneva-Atanasova K, Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021 Mar 9;372:n579. PubMed: https://pubmed.gov/33687922. Full-text: https://doi.org/10.1136/bmj.n579
Is B.1.1.7 more lethal? In this large study from the UK, mortality hazard ratio associated with infection with “B.1.1.7” (S gene negative) was 1.64 (95% CI: 1.32 to 2.04) in the community, compared with previously circulating variants. However, even though the authors controlled for some biases, their matched control approach bears several limitations. Let’s take it as a first hint, but not as proof.
16 March
Garcia-Beltran WF, Lam EC, Denis KS, et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell March 12, 2021. https://www.cell.com/cell/fulltext/S0092-8674(21)00298-1
Among 99 individuals who received one or two doses of mRNA vaccines, most individuals receiving a single dose did not raise sufficient antibody titers to provide detectable cross neutralization to B.1.351. This supports the importance of 2-dose regimens to acheive protective titers.
Altman DM, Boyton RJ, Beale R. Immunity to SARS-CoV-2 variants of concern. Science 12 Mar 2021: Vol. 371, Issue 6534, pp. 1103-1104. https://science.sciencemag.org/content/371/6534/1103
Brilliant brief review about not only immunity to variants but also on methodological issues. Assessment of variants on neutralization are complicated by the variability of pseudotype assays used.
Graphic
Li Y, Ma ML, Lei Q, et al. Linear epitope landscape of the SARS-CoV-2 Spike protein constructed from 1,051 COVID-19 patients. Cell Reports March 12, 2021. DOI:https://doi.org/10.1016/j.celrep.2021.108915
What part of the spike protein is highly immunogeneic? By analyzing the serum IgG response of 1051 COVID-19 patients with a peptide microarray, the authors built a comprehensive epitope landscape that covers the entire sequence of the SARS-CoV-2 spike protein. A set of 16 highly immunogenic epitopes outside of the RBD region were identified. The antibody responses against several epitopes are associated with severity. Little neutralization activity was observed for the antibodies against the highly immunogenic epitopes.
14 March
Grint DJ, Wing K, Williamson E, et al. Case fatality risk of the SARS-CoV-2 variant of concern B.1.1.7 in England. MedRxiv 2021, posted 8 March. Full-text: https://doi.org/10.1101/2021.03.04.21252528
The authors draw data from a research platform that covers 40% of England’s population registered with a general practitioner. B.1.1.7 status was known for 184,786 people. The B.1.1.7 group was younger with a lower proportion of older cases (80+: 0.9% VOC vs. 1.6% non-B.1.1.7 cases), with fewer Comorbidities (2+ Comorbidities: 2.9% vs. 3.8%). After controlling for Comorbidities, age, week, region & other sociodemographics, the authors found an increased risk of death for B.1.1.7 compared with non-B.1.1.7 cases (HR: 1.67; 95% CI: 1.34 – 2.09; P < 0.0001).
Tablizo FA, Kim KM, Lapid CM, et al. Genome sequencing and analysis of an emergent SARS-CoV-2 variant characterized by multiple spike protein mutations detected from the Central Visayas Region of the Philippines. medRxiv 2021, posted 6 March. Full-text: https://doi.org/10.1101/2021.03.03.21252812
The authors describe the emergence of a new SARS-CoV-2 lineage, mainly from the Central Visayas region of the Philippines: 13 lineage-defining mutations, including the co-occurrence of the E484K, N501Y, and P681H mutations at the spike protein region, as well as three additional radical amino acid replacements towards the C-terminal end of the said protein. A three amino acid deletion at positions 141 to 143 (LGV141_143del) in the spike protein is reminiscent of a region preceding the 144Y deletion found in the B.1.1.7 variant. See also the ‘P.3’ proposition by Andrew Rambaut: https://github.com/cov-lineages/pango-designation/issues/27.
13 March
Paper of the Day
Collier DA, De Marco A, Ferreira IA, et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature March 11, 2021. https://www.nature.com/articles/s41586-021-03412-7
A pseudovirus bearing S protein with the full set of mutations present in the B.1.1.7 variant did result in a small reduction in neutralization by sera from vaccinees that was more marked following the first dose vs the second dose. Worryingly, Dami A. Collier and colleagues measured further reduction in neutralization titers by vaccine sera when E484K was present alongside the B.1.1.7 S mutations. They conclude that “E484K emergence on a B.1.1.7 background represents a threat to the vaccine BNT162b”.
Borges V, Sousa C, Menezes L, et al. Tracking SARS-CoV-2 lineage B.1.1.7 dissemination: insights from nationwide spike gene target failure (SGTF) and spike gene late detection (SGTL) data, Portugal, week 49 2020 to week 3 2021. Eurosurveillance Mar 11, 2021, Article Volume 26, Issue 10. https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2021.26.10.2100130
Dissemination of the B.1.1.7 lineage in Portugal. Both SGTF and SGTL (a proxy for monitoring trends of B.1.1.7) samples had significantly lower median Ct values of N and ORF1ab gene targets (ca 3.5 and 1.8 Ct units, respectively) compared with samples where the S gene was unbiasedly detected.
11 March
Paper of the Day
Tegally H, Wilkinson E, Giovanetti M. et al. Emergence of a SARS-CoV-2 variant of concern with mutations in spike glycoprotein. Nature March 9, 2021. https://www.nature.com/articles/s41586-021-03402-9_reference.pdf
The future “reference” paper on the detection of B.1.351 (or 501Y.V2), characterized by eight lineage-defining mutations in the spike protein, including three at important residues in the receptor-binding domain (K417N, E484K and N501Y). This lineage was identified in South Africa after the first epidemic wave in a severely affected metropolitan area, and spread rapidly. The genomic data, showing the rapid expansion and displacement of other lineages in multiple regions, suggest that this lineage is associated with a selection advantage, most plausibly as a result of increased transmissibility or immune escape. Nevertheless, congrats to co-author Wolfgang Preiser!
10 March
Paper of the Day
Wang P, Nair MS, Liu L, et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature March 8, 2021. https://www.nature.com/articles/s41586-021-03398-2
For younger readers: once upon a time, in the stone age (c. 1996), David Ho from the Aaron Diamond Center was deemed “man of the year” (Time Magazine), after explaining the dynamics of HIV replication to the world. Today he explains why B.1.351 is so worrisome. While B.1.1.7 is refractory to neutralization by many mAbs but not more resistant to convalescent plasma (CP) or vaccinee sera (VS), B.1.351 is not only refractory to neutralization by almost all mAbs but also by CP (9.4 fold) and VS (10.3-12.4 fold). SARS-CoV-2 “is traveling in a direction that could ultimately lead to escape from our current therapeutic and prophylactic interventions directed to the viral spike”.
9 March
Alexandre G, Bosetti P, Feri A, et al. Early assessment of diffusion and possible expansion of SARS-CoV-2 Lineage 20I/501Y.V1 (B.1.1.7, variant of concern 202012/01) in France, January to March 2021. Euro Surveill. 2021;26(9):pii=2100133. https://doi.org/10.2807/1560-7917.ES.2021.26.9.2100133
Spread of B.1.1.7. in France, in January. The authors estimate the population-level effective reproduction number will be respectively 39% (95%: 33–45%) and 56% (95%: 50–62%) higher on 1 March and 1 April 2021 than what would be expected if only the classical lineages were circulating.
8 March
Firestone MJ, Lorentz AJ, Meyer S, et al. First Identified Cases of SARS-CoV-2 Variant P.1 in the United States — Minnesota, January 2021. MMWR Morb Mortal Wkly Rep. ePub: 3 March 2021. https://www.cdc.gov/mmwr/volumes/70/wr/mm7010e1.htm?s_cid=mm7010e1_x
The arrival of P.1 in the US. Both guys had returned from southeastern Brazil. According to the authors, this “underscores the importance of community prevention strategies to slow transmission of SARS-CoV-2 including use of well-fitting masks, physical distancing, washing hands, quarantine, testing of persons who have had contact with a person with laboratory-confirmed COVID-19, isolating persons with symptoms of COVID-19 or with diagnosed COVID-19 and” (drumroll, please) “adhering to CDC recommendations to delay travel”. Maybe it’s not the best idea to vacation in Brazil (or anywhere!) right now.
Ojelade M, Rodriguez A, Gonzalez D, et al. Travel from the United Kingdom to the United States by a Symptomatic Patient Infected with the SARS-CoV-2 B.1.1.7 Variant — Texas, January 2021. MMWR Morb Mortal Wkly Rep. ePub: 3 March 2021. https://www.cdc.gov/mmwr/volumes/70/wr/mm7010e2.htm?s_cid=mm7010e2_w
Another jerk who traveled from UK to US after experiencing COVID-19–compatible symptoms, infected with the B.1.1.7 variant. “Persons should not travel if they are experiencing symptoms compatible with COVID-19 or if they have received a positive SARS-CoV-2 test result and have not met criteria to discontinue isolation, have had close contact with a person with suspected or confirmed COVID-19 and have not subsequently met criteria to end quarantine, or have a pending SARS-CoV-2 viral test result”. Questions?
Fujino T, Nomoto H, Kutsuna S, Ujiie M, Suzuki T, Sato R, et al. Novel SARS-CoV-2 variant identified in travelers from Brazil to Japan. Emerg Infect Dis. 2021 Apr [date cited]. https://doi.org/10.3201/eid2704.210138
A family of four, traveling in early January to Tokyo, Japan, from Amazonas state in Brazil via Istanbul, Turkey (by the way, wouldn’t it have been shorter heading west?). Souvenir: A new lineage, resembling P.1, but with some interesting new mutations. Cringe.
Maggi F, Novazzi F, Genoni A, Baj A, Spezia PG, Focosi D, et al. Imported SARS-CoV-2 variant P.1 detected in traveler returning from Brazil to Italy. Emerg Infect Dis. 2021 Apr [date cited]. https://doi.org/10.3201/eid2704.210183
A family of three, flying from São Paulo, Brazil, via Madrid, Spain, to Milan (Malpensa Airport) in Italy, in mid-January. In the luggage: P.1.
7 March
Paper of the Day
De Souza WM, Amorm MR, Sesti-Costa R, et al. Levels of SARS-CoV-2 Lineage P.1 Neutralization by Antibodies Elicited after Natural Infection and Vaccination. Lancet Preprints 2021, posted 1 March. Full-text: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3793486
William de Souza and colleagues isolated two P.1-containing specimens from nasopharyngeal and bronchoalveolar lavage samples of patients in Manaus, Brazil. They found that the immune plasma of COVID-19 convalescent blood donors had 6-fold less neutralizing capacity against the P.1 than against the B lineage. Moreover, five months after booster immunization with the Chinese CoronaVac vaccine, plasma from vaccinated individuals failed to efficiently neutralize P.1 lineage isolates.
Faria NR, Mellan TA, Whittaker C, et al. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. MedRxiv 2021, posted 3 March. Full-text: https://doi.org/10.1101/2021.02.26.21252554
Using a combination of genomic and epidemiological data, Nuno Faria and colleagues characterize the emergence and characteristics of P.1 that acquired 17 mutations, including the trio in the spike protein (K417T, E484K and N501Y) associated with increased binding to the human ACE2 receptor. The authors show that P.1 emerged around early November 2020. They estimate that P.1 could be 1.4–2.2 times more transmissible and able to evade 25-61% of protective immunity elicited by previous infection with non-P.1 lineages.
Wang P, Wang M, Yu J, et al. Increased Resistance of SARS-CoV-2 Variant P.1 to Antibody Neutralization. bioRxiv 2021, posted 2 March. Full-text: https://doi.org/10.1101/2021.03.01.433466
David D. Ho, Pengfei Wang and colleagues report that P.1 is not only refractory to multiple neutralizing monoclonal antibodies, but also more resistant to neutralization by convalescent plasma (6.5-fold) and vaccinee sera (2.2-2.8-fold).
Cele S, Gazy I, Jackson L, et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. MedRxiv 2021, posted 27 February. Full-text: https://doi.org/10.1101/2021.01.26.21250224
We already knew that people previously infected with the non-B.1.351 variant don’t neutralize B.1.351 very effectively. Now Alex Sigal, Tulio de Oliveira, Sandile Cele and colleagues show that people infected with B.1.351 can neutralize both B.1.351 and (to a slightly lesser extent) ‘regular’ non-B.1.351 viruses (Cele 2021). If these data are confirmed, a variant B.1.351-targeted booster vaccine could be a solution for countries where B.1.351 is the dominant strain.
Starr TN, Greaney AJ, Dingens AS, Bloom JD. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. bioRxiv 2021, posted 21 February. Full-text: https://doi.org/10.1101/2021.02.17.431683
Jesse Bloom, Tyler Starr and colleagues completely map all mutations to the SARS-CoV-2 spike receptor binding domain (RBD) that escape binding by LY-CoV555 (bamlanivimab, a monoclonal antibody manufactured by Lilly), and its cocktail combination with LY-CoV016. Individual mutations that escape binding are present in B.1.351 and P.1 (E484K escapes LY-CoV555, K417N/T escape LY-CoV016). Additionally, the L452R mutation in the B.1.429 lineage escapes LY-CoV555.
6 March
Paper of the Day
Shen X, Tang H, McDanal C, et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral Spike vaccines. Cell Host Microbe March 03, 2021. https://www.cell.com/cell-host-microbe/fulltext/S1931-3128(21)00102-5
Good news. Using a lentivirus-based pseudovirus assay, Xiaoying Shen and colleagues from Duke, Durham, US, show that B.1.1.7 is probably not a neutralization escape variant of concern for COVID-19 vaccines. Moreover, B.1.1.7 is unlikely to increase the risk of SARS-CoV-2 re-infection.
Wibmer CK, Ayres F, Hermanus T, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nature Medicine 02 March 2021. https://www.nature.com/articles/s41591-021-01285-x
B.1.351 is a bigger problem. This study (for months available as a pre-print only, now as a beautiful paper in Nature Medicine) shows that this lineage completely escapes three classes of therapeutically relevant antibodies. The B.1.351 pseudovirus also exhibited substantial to complete escape from neutralization, but not binding, by convalescent plasma. The overwhelming majority of monoclonal antibodies already on the path to licensure target residues K417 or E484 and are therefore likely to be futile against this variant.
5 March
Paper of the Day
Davies NG, Abbott S, Barnard RS, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 03 Mar 2021: eabg3055. Full-text: https://science.sciencemag.org/content/early/2021/03/03/science.abg3055
Difficult months ahead: the B.1.1.7 variant emerged in southeast England in November 2020 and is rapidly spreading toward dominance. In this brilliant article, Nicholas Davies and colleagues from London estimate that this variant has a 43–90% (range of 95% credible intervals 38–130%) higher reproduction number than pre-existing variants. A fitted two-strain dynamic transmission model shows that B.1.1.7. will lead to large resurgences of COVID-19 cases. Without stringent control measures, including limited closure of educational institutions and a greatly accelerated vaccine roll-out, COVID-19 hospitalizations and deaths across England in 2021 will exceed those in 2020.
4 March
Plante JA, Mitchell BM, Plante KS, et al. The Variant Gambit: COVID’s Next Move. Cell Host Microbe March 01, 2021. https://www.cell.com/cell-host-microbe/fulltext/S1931-3128(21)00099-8
Nice review that outlines factors driving SARS-CoV-2 variant evolution, explores the potential impact of specific mutations and examines the risk of further mutations. Jessica A. Plante and colleagues from the World Reference Center for Emerging Viruses and Arboviruses consider also the experimental studies needed to understand the threat these variants pose.
28 February
Paper of the Day
WHO 20210223. Weekly epidemiological update – 23 February 2021. WHO 2021, published 23 February. Full-text: https://www.who.int/publications/m/item/weekly-epidemiological-update—23-february-2021
If you are interested in variants, read the Special Focus: Update on SARS-CoV-2 Variants of Concern on page 7 to 13 with an excellent table on the current information we have on B.1.1.7, B.1.351 and P.1.
Naveca F, Nascimento V, Souza V, et al. COVID-19 epidemic in the Brazilian state of Amazonas was driven by long-term persistence of endemic SARS-CoV-2 lineages and the recent emergence of the new Variant of Concern P.1. Research Square 2021, posted 25 February. Full-text: https://doi.org/10.21203/rs.3.rs-275494/v1
The second wave in the Northern Brazilian state of Amazonas coincides with the emergence of the P.1 variant in late November 2020. P.1 replaced the parental lineage in less than two months. The authors report that successive lineage replacements in Amazonas were driven by a complex combination of variable levels of social distancing measures and the emergence of a more transmissible VOC P.1 virus. They provide insights to understanding the mechanisms that underlie COVID-19 waves and the risk of disseminating P.1 in Brazil and potentially worldwide.
Faria NR, Mellan TA, Whittaker C, et al. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus. GitHub 2021, posted 27 February. Full-text: https://github.com/CADDE-CENTRE/Novel-SARS-CoV-2-P1-Lineage-in-Brazil/blob/main/manuscript/FINAL_P1_MANUSCRIPT_25-02-2021_combined.pdf
Using a combination of genomic and epidemiological data, Nuno Faria and colleagues characterize the emergence and characteristics of P.1 that acquired 17 mutations, including the trio in the spike protein (K417T, E484K and N501Y) associated with increased binding to the human ACE2 receptor. The authors show that P.1 emerged around early November 2020. They estimate that P.1 could be 1.4–2.2 times more transmissible and able to evade 25-61% of protective immunity elicited by previous infection with non-P.1 lineages. These data need to be confirmed by further studies.
25 February
Paper of the Day
Zhou D, Dejnirattisai W, Supasa P, et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine induced sera. Cell February 23, 2021. Full-text: https://www.cell.com/action/showPdf?pii=S0092-8674%2821%2900226-9
The new variants have multiple changes in the immunodominant spike protein which facilitates viral cell entry via the ACE receptor. Mutations in the receptor recognition site on the spike are of great concern due to their potential for immune escape. Daming Zhou and colleagues from Oxford, UK describe a structure-function analysis of B.1.351 using a large cohort of convalescent and vaccinee serum samples. The receptor binding domain mutations provide tighter ACE2 binding and widespread escape from monoclonal antibody neutralization largely driven by E484K although K417N and N501Y act together against some important antibody classes. The neutralization titer for B.1.351 reduced 8 to 9-fold for both the Pfizer and AstraZeneca vaccinees. E484K, K417N and N501Y caused widespread escape from monoclonal antibodies. However, let‘s keep in mind that even if antibody responses to the new variants are not able to prevent infection, they may moderate severity. Moreover, T cell responses to spike may not be disrupted by the mutational changes described here.
Li Q, Nie J, Wu J. No higher infectivity but immune escape of SARS-CoV-2 501Y.V2 variants. Cell February 23, 2021. https://www.cell.com/action/showPdf?pii=S0092-8674%2821%2900231-2
More experiments on B.1.351 (also known as 501Y.V2). These variants DO NOT confer increased infectivity in multiple cell types except for murine (not human!) ACE2-overexpressing cells, where a substantial increase in infectivity was observed. As seen in the other paper, the susceptibility of the variants to neutralizing monoclonal antibodies was substantially diminished, and the neutralization ability of the sera from convalescent patients and immunized mice was also reduced. The neutralization resistance was mainly caused by E484K and N501Y mutations in the receptor-binding domain of Spike.
Vasques Nonaka CK, Franco MM, Gräf T, et al. Genomic evidence of SARS-CoV-2 reinfection involving E484K spike mutation, Brazil. Emerg Infect Dis. February 19, 2021. https://wwwnc.cdc.gov/eid/article/27/5/21-0191_article
A case of reinfection from distinct virus lineages in Brazil harboring the E484K mutation, a variant associated with escape from neutralizing antibodies (see above). Both episodes were considered to be mild.
20 February
Liu Y, Liu J, Xia H, et al. Neutralizing Activity of BNT162b2-Elicited Serum – Preliminary Report. N Engl J Med. 2021 Feb 17. PubMed: https://pubmed.gov/33596352. Full-text: https://doi.org/10.1056/NEJMc2102017
Some more in vitro data on vaccine efficacy in SARS-CoV-2 variants. The authors produced different recombinant viruses, among them one with all the mutations found in the S gene in the B.1.351 lineage. After a single shot of BNT162b2 from Pfizer/BioNTech, neutralization of the B.1.351-spike virus was weaker by approximately two thirds. However, according to Yang Liu and colleagues, it remains unclear what this reduction means in terms of protection.
Wu K, Werner AP, Koch M, et al. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine – Preliminary Report. N Engl J Med. 2021 Feb 17. PubMed: https://pubmed.gov/33596346. Full-text: https://doi.org/10.1056/NEJMc2102179
Same with Moderna’s vaccine. In vitro reductions by a factor of 2,7-6,4 in titers of neutralizing antibodies against the partial or full panel of mutations. Again, protection against B.1.351 remains to be determined.
Twitter: Some more in vitro data on vaccine efficacy in SARS-CoV-2 variants (in particular B.1.351), after BNT162b2 from Pfizer/BioNTech https://doi.org/10.1056/NEJMc2102017 and Moderna https://doi.org/10.1056/NEJMc2102179
19 February
Paper of the Day
Garcia-Beltran WF, Lam EC, St. Denis K, et al. Circulating SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. medRxiv 2021, posted 18 February. Full-text: https://doi.org/10.1101/2021.02.14.21251704
While many strains, such as B.1.1.7, B.1.1.298, or B.1.429, continue to be potently neutralized despite the presence of individual receptor-binding domain (RBD) mutations, other circulating SARS-CoV-2 variants escape vaccine-induced humoral immunity. The P.2 variant, which contains an E484K mutation within the RBD region, was capable of significantly reducing the neutralization potency of fully vaccinated individuals. Similarly, the P.1 strain, which has three RBD mutations, more effectively escaped neutralization. Finally, B.1.351 variants exhibited remarkable resistance to neutralization, largely due to three mutations in RBD but with a measurable contribution from non-RBD mutations. The magnitude of the effect is such that B.1.351 strains escape neutralizing vaccine responses like SARS-CoV-1 (SARS 2002/2003) and bat-derived WIV1-CoV, suggesting that a relatively small number of mutations can mediate potent escape from vaccine responses. Alejandro Balazs, Wilfredo F. Garcia-Beltran and colleagues emphasize the need to develop broadly protective interventions against the evolving pandemic.
Resende PC, Delatorre D, Gräf T, et al. Evolutionary Dynamics and Dissemination Pattern of the SARS-CoV-2 Lineage B.1.1.33 During the Early Pandemic Phase in Brazil. Front. Microbiol 2021, 17 February. Full-text: https://www.frontiersin.org/articles/10.3389/fmicb.2020.615280/
Paula Cristina Resende et al. investigated the origin of the major and most widely disseminated SARS-CoV-2 Brazilian lineage B.1.1.33 that evolved from an ancestral clade, here designated B.1.1.33-like. The B.1.1.33-like lineage may have been introduced from Europe or may have arisen in Brazil in early February 2020 and a few weeks later gave origin to the lineage B.1.1.33.
18 February
Hodcroft EB, Domman DB, Snyder DJ, et al. Emergence in late 2020 of multiple lineages of SARS-CoV-2 Spike protein variants affecting amino acid position 677. medRxiv 2021, posted 14 January. Full-text: https://doi.org/10.1101/2021.02.12.21251658
Emma Hodcroft et al. describe 7 newly identified coronavirus variants in the US with a mutation in spike position 677 (also named after birds, Mockingbird to Yellowhammer). The authors promise to keep an eye on S:677 polymorphisms for effects on proteolytic processing, cell tropism, and transmissibility.
Hoffmann M, Arora P, Groß R, et al. SARS-CoV-2 variants B.1.351 and B.1.1.248: Escape from therapeutic antibodies and antibodies induced by infection and vaccination. medRxiv 2021, posted 11 February. Full-text: https://doi.org/10.1101/2021.02.11.430787
Stefan Pöhlmann, Markus Hoffmann and colleagues show that B1351 (first detected in South Africa) and P1 (alias B11248, first detected in Brazil) were partially (casirivimab, in REGN-COV2, Regeneron) or fully (bamlanivimab, Lilly) resistant to monoclonal antibodies and was less efficiently inhibited by serum/plasma from convalescent individuals or those vaccinated with the Pfizer-BioNTech vaccine.
15 February
Paper of the Day
NERVTAG 20210211. Update note on B.1.1.7 severity. New and Emerging Respiratory Virus Threats Advisory Group (NERVTAG) 2021, published 12 February. Full-text: https://www.gov.uk/government/publications/nervtag-update-note-on-b117-severity-11-february-2021
It is now likely that infection with B.1.1.7 is associated with an increased risk of hospitalization and death compared to infection with previously circulating viruses.
Variants – 13 February update. COVID Reference 2021, published 13 February. Full-text: https://covidreference.com/variants | See also the Comparison document with new content added between 7 and 13 February: http://www.bsk1.com/CR_Variants_Update.html
In most of continental Europe and the US, Easter 2021 (4 April) may be recalled later as a B.1.1.7 Easter.
14 February
Fontanet A, Autran B, Lina B, et al. SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet 2021, published 11 February. Full-text: https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(21)00370-6/fulltext
Arnaud Fontanet from the Pasteur Institute in Paris outlines the priorities to address in an environment with new SARS-CoV-2 variants:
- Continue to suppress and push to eliminate SARS-CoV-2 while rolling out COVID-19 vaccines
- Improve surveillance of SARS-CoV-2 variants through global sequencing and sharing of variant-specific PCR primers
- Create a central repository of samples of sera and cells from individuals with past infection or past immunization with available COVID-19 vaccines for sero-neutralization and cellular immunity functional testing against newly discovered variants
- Produce COVID-19 vaccines reactively and adapt them to newly emerging lineages
- Ensure global access, availability, and affordability of COVID-19 vaccines to ensure no countries are left behind
13 February
Zhang W, Davis BD, Chen SS, Sincuir Martinez JM, Plummer JT, Vail E. Emergence of a Novel SARS-CoV-2 Variant in Southern California. JAMA. 2021 Feb 11. PubMed: https://pubmed.gov/33571356. Full-text: https://doi.org/10.1001/jama.2021.1612
In California, the proportion of SARS-CoV-2 cases associated with the CAL.20C variant rose from 3,8% to 25% between mid-November and late December. By then, it accounted for 24% of samples in one study, and 36,4% (66/181) of samples in a local Los Angeles cohort. The emerging predominance of this strain is temporally related to the time of onset of the current spike in SARS-CoV-2 infections in Southern California. CAL.20C is defined by mutations in the S protein (L452R, S13I, W152C) and in the ORF1a (I4205V) and ORF1b protein (D1183Y).
9 February
Althaus C, et al. Transmission of SARS-CoV-2 variants in Switzerland. Institute of Social and Preventive Medicine (ISPM), University of Bern 2021, reported 5 February. Full-text: https://ispmbern.github.io/covid-19/variants/
For 5 February, the authors estimated the proportion of SARS-CoV-2 variants (501Y, B.1.1.7) to have reached 67% in Geneva and 35% in Zurich. They also estimated the increase in transmissibility slightly above 50%.
8 February
Paper of the Day
Washington NL, Gangavarapu K, Zeller M, et al. Genomic epidemiology identifies emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. medRxiv 2021, posted 7 February. Full-text: https://www.medrxiv.org/content/10.1101/2021.02.06.21251159v1
Kristian Andersen, Nicole L. Washington, Karthik Gangavarapu and colleagues from Helix and the Scripps Research Institute, La Jolla, sequenced 212 B117 genomes collected in the U.S. from December 2020 to January 2021. They found a doubling rate of a little over a week and an increased transmission rate of 35-45%. The authors show that the U.S. is on a similar trajectory as other countries where B117 rapidly became the dominant SARS-CoV-2 variant and warn that immediate and decisive action to minimize COVID-19 morbidity and mortality. Authorities in Germany, France, Italy, Spain and other countries should listen too.
2 February
With viruses, some mutations emerge while others recede. Rarely does one or more mutations confer a “selective advantage” to a new variant, for example enhanced transmissibility. Such variants can then become the new dominant virus.
Introduction
Over the last two months, several new SARS-CoV-2 variants have been described that are more transmissible, may escape both natural and vaccine-induced immunity and could impact COVID-19 morbidity and mortality. It is too early to assert that these variants will create a new pandemic within the pandemic, however, in countries like England, South Africa, Brazil, Ireland, Portugal and Israel, they may have modified the dynamic of the latest outbreaks for the worse. More transmissible SARS-CoV-2 variants will replace older variants – everywhere! Countries where the prevalence of these new variants is still low should anticipate rapid spread within the next weeks and months and plan ahead accordingly, ie closing/restricting borders, etc.
The current trio infernale:
- B117 (first described in England; Rambaut 2020)
- B1351 (first described in South Africa; Tegally 2020)
- P1 (first described in Brazil; Faria 2021)
